
Blautia sp. An249
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Blautia; unclassified Blautia
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
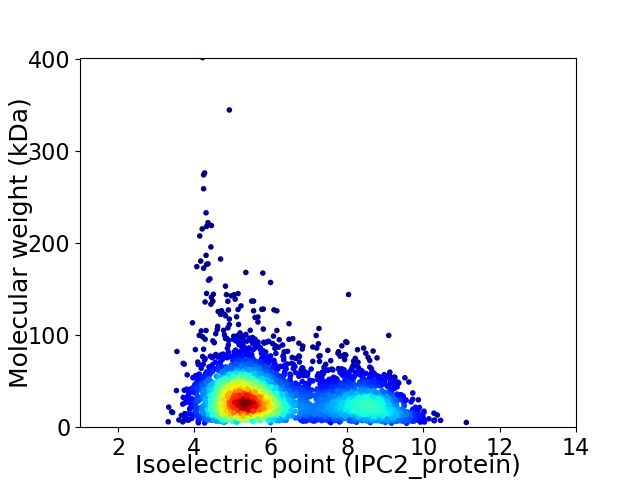
Virtual 2D-PAGE plot for 3735 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
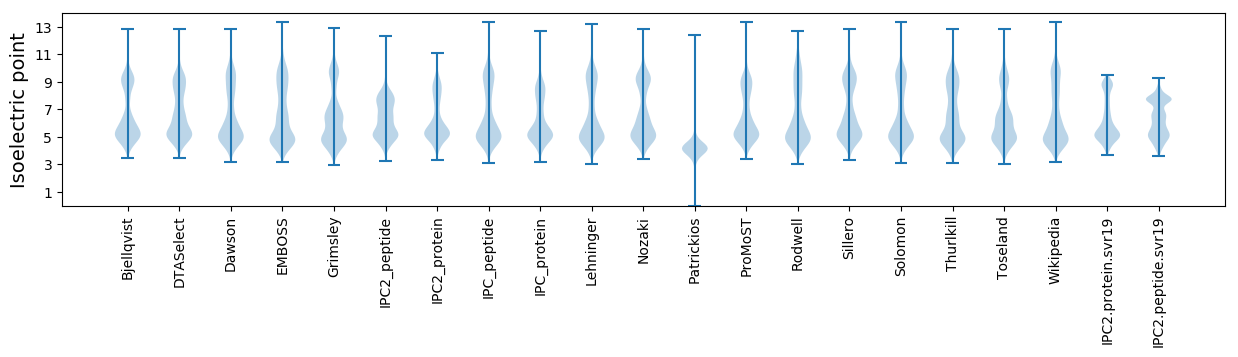
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4F6F3|A0A1Y4F6F3_9FIRM TM1586_NiRdase domain-containing protein OS=Blautia sp. An249 OX=1965603 GN=B5F53_09470 PE=4 SV=1
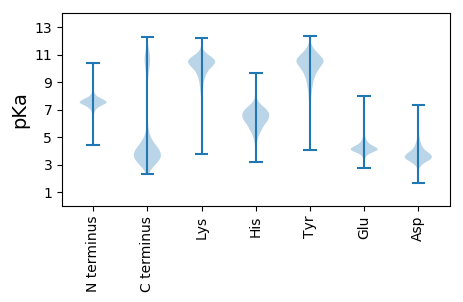
MM1 pKa = 7.42SRR3 pKa = 11.84KK4 pKa = 9.38HH5 pKa = 6.18KK6 pKa = 10.74NLVILATVLSAACIFSACGKK26 pKa = 10.06DD27 pKa = 4.04KK28 pKa = 10.91EE29 pKa = 4.48DD30 pKa = 3.36QVVVAEE36 pKa = 4.94APTPTPTVAPTITPVPSVAATAQKK60 pKa = 8.9VTYY63 pKa = 8.48TSKK66 pKa = 10.81DD67 pKa = 3.35SSVSIVLPDD76 pKa = 3.66NTWKK80 pKa = 8.81NTSDD84 pKa = 3.3NGKK87 pKa = 10.49SIIFNSDD94 pKa = 2.66TQGNISITYY103 pKa = 10.1ANTDD107 pKa = 2.92ADD109 pKa = 4.17LAKK112 pKa = 10.81VQIPSSKK119 pKa = 9.94EE120 pKa = 3.42LAGSITAGTSLTEE133 pKa = 4.06GTDD136 pKa = 4.15FDD138 pKa = 4.14ILDD141 pKa = 3.74YY142 pKa = 10.51TATEE146 pKa = 3.87MDD148 pKa = 3.43GVDD151 pKa = 3.27VYY153 pKa = 11.18RR154 pKa = 11.84YY155 pKa = 6.55TVKK158 pKa = 10.76AADD161 pKa = 3.47ASKK164 pKa = 9.96MNGYY168 pKa = 10.79AYY170 pKa = 9.76IEE172 pKa = 4.2NYY174 pKa = 10.34YY175 pKa = 9.21LTDD178 pKa = 3.67GSEE181 pKa = 4.69LYY183 pKa = 10.52QISGSLKK190 pKa = 10.28LQDD193 pKa = 3.69PTSLNTMKK201 pKa = 10.62EE202 pKa = 3.96AVTSFRR208 pKa = 11.84ILKK211 pKa = 10.1ADD213 pKa = 3.79SPLKK217 pKa = 10.61AAADD221 pKa = 4.47TIASQNSSSATPSVTPSVTPAASGDD246 pKa = 3.79SDD248 pKa = 4.52NSDD251 pKa = 3.52SSDD254 pKa = 4.11DD255 pKa = 3.78SDD257 pKa = 4.25TNTSSTSDD265 pKa = 3.3GSSSDD270 pKa = 2.98SSYY273 pKa = 11.77DD274 pKa = 3.63DD275 pKa = 4.03SSSSGDD281 pKa = 3.47DD282 pKa = 3.3SGSGNSGQSSEE293 pKa = 4.71DD294 pKa = 2.71GWYY297 pKa = 10.42DD298 pKa = 3.18GDD300 pKa = 4.06TFISYY305 pKa = 11.09DD306 pKa = 3.82DD307 pKa = 5.31GYY309 pKa = 11.46FDD311 pKa = 6.73DD312 pKa = 5.4DD313 pKa = 6.31GYY315 pKa = 9.33WHH317 pKa = 7.73WYY319 pKa = 9.76GDD321 pKa = 3.81DD322 pKa = 3.96SGDD325 pKa = 5.07DD326 pKa = 3.74GSGDD330 pKa = 4.06DD331 pKa = 4.76GSDD334 pKa = 4.28DD335 pKa = 3.87GSSDD339 pKa = 4.85DD340 pKa = 4.73GSGDD344 pKa = 3.95DD345 pKa = 5.63GSGSDD350 pKa = 4.92DD351 pKa = 4.49GGWYY355 pKa = 10.01DD356 pKa = 3.78NNGNYY361 pKa = 9.72FSYY364 pKa = 11.35NDD366 pKa = 4.75GYY368 pKa = 11.39FDD370 pKa = 6.87DD371 pKa = 5.77DD372 pKa = 6.31GVWQWYY378 pKa = 8.98
MM1 pKa = 7.42SRR3 pKa = 11.84KK4 pKa = 9.38HH5 pKa = 6.18KK6 pKa = 10.74NLVILATVLSAACIFSACGKK26 pKa = 10.06DD27 pKa = 4.04KK28 pKa = 10.91EE29 pKa = 4.48DD30 pKa = 3.36QVVVAEE36 pKa = 4.94APTPTPTVAPTITPVPSVAATAQKK60 pKa = 8.9VTYY63 pKa = 8.48TSKK66 pKa = 10.81DD67 pKa = 3.35SSVSIVLPDD76 pKa = 3.66NTWKK80 pKa = 8.81NTSDD84 pKa = 3.3NGKK87 pKa = 10.49SIIFNSDD94 pKa = 2.66TQGNISITYY103 pKa = 10.1ANTDD107 pKa = 2.92ADD109 pKa = 4.17LAKK112 pKa = 10.81VQIPSSKK119 pKa = 9.94EE120 pKa = 3.42LAGSITAGTSLTEE133 pKa = 4.06GTDD136 pKa = 4.15FDD138 pKa = 4.14ILDD141 pKa = 3.74YY142 pKa = 10.51TATEE146 pKa = 3.87MDD148 pKa = 3.43GVDD151 pKa = 3.27VYY153 pKa = 11.18RR154 pKa = 11.84YY155 pKa = 6.55TVKK158 pKa = 10.76AADD161 pKa = 3.47ASKK164 pKa = 9.96MNGYY168 pKa = 10.79AYY170 pKa = 9.76IEE172 pKa = 4.2NYY174 pKa = 10.34YY175 pKa = 9.21LTDD178 pKa = 3.67GSEE181 pKa = 4.69LYY183 pKa = 10.52QISGSLKK190 pKa = 10.28LQDD193 pKa = 3.69PTSLNTMKK201 pKa = 10.62EE202 pKa = 3.96AVTSFRR208 pKa = 11.84ILKK211 pKa = 10.1ADD213 pKa = 3.79SPLKK217 pKa = 10.61AAADD221 pKa = 4.47TIASQNSSSATPSVTPSVTPAASGDD246 pKa = 3.79SDD248 pKa = 4.52NSDD251 pKa = 3.52SSDD254 pKa = 4.11DD255 pKa = 3.78SDD257 pKa = 4.25TNTSSTSDD265 pKa = 3.3GSSSDD270 pKa = 2.98SSYY273 pKa = 11.77DD274 pKa = 3.63DD275 pKa = 4.03SSSSGDD281 pKa = 3.47DD282 pKa = 3.3SGSGNSGQSSEE293 pKa = 4.71DD294 pKa = 2.71GWYY297 pKa = 10.42DD298 pKa = 3.18GDD300 pKa = 4.06TFISYY305 pKa = 11.09DD306 pKa = 3.82DD307 pKa = 5.31GYY309 pKa = 11.46FDD311 pKa = 6.73DD312 pKa = 5.4DD313 pKa = 6.31GYY315 pKa = 9.33WHH317 pKa = 7.73WYY319 pKa = 9.76GDD321 pKa = 3.81DD322 pKa = 3.96SGDD325 pKa = 5.07DD326 pKa = 3.74GSGDD330 pKa = 4.06DD331 pKa = 4.76GSDD334 pKa = 4.28DD335 pKa = 3.87GSSDD339 pKa = 4.85DD340 pKa = 4.73GSGDD344 pKa = 3.95DD345 pKa = 5.63GSGSDD350 pKa = 4.92DD351 pKa = 4.49GGWYY355 pKa = 10.01DD356 pKa = 3.78NNGNYY361 pKa = 9.72FSYY364 pKa = 11.35NDD366 pKa = 4.75GYY368 pKa = 11.39FDD370 pKa = 6.87DD371 pKa = 5.77DD372 pKa = 6.31GVWQWYY378 pKa = 8.98
Molecular weight: 39.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4F598|A0A1Y4F598_9FIRM Inosine monophosphate cyclohydrolase OS=Blautia sp. An249 OX=1965603 GN=B5F53_09430 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.89TKK25 pKa = 10.34GGRR28 pKa = 11.84KK29 pKa = 8.67VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.89TKK25 pKa = 10.34GGRR28 pKa = 11.84KK29 pKa = 8.67VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1167479 |
37 |
3650 |
312.6 |
35.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.321 ± 0.044 | 1.516 ± 0.018 |
5.229 ± 0.037 | 8.243 ± 0.045 |
4.037 ± 0.026 | 7.262 ± 0.038 |
1.673 ± 0.016 | 7.037 ± 0.038 |
6.852 ± 0.039 | 9.16 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.99 ± 0.022 | 3.987 ± 0.025 |
3.409 ± 0.022 | 3.654 ± 0.021 |
4.639 ± 0.03 | 5.824 ± 0.038 |
5.426 ± 0.039 | 6.638 ± 0.036 |
0.971 ± 0.014 | 4.132 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
