
uncultured marine virus
Taxonomy: Viruses; environmental samples
Average proteome isoelectric point is 8.28
Get precalculated fractions of proteins
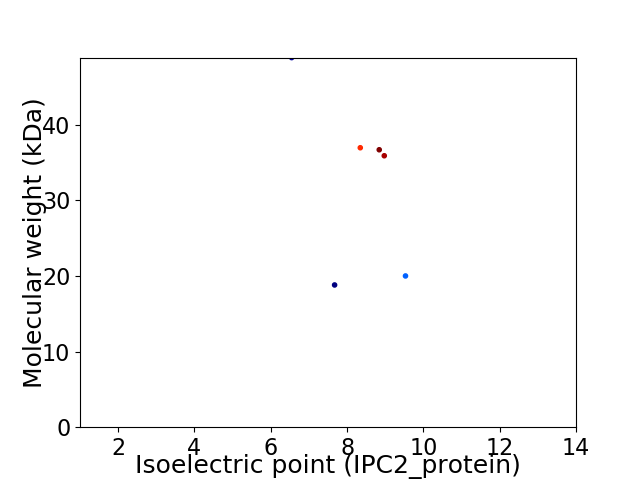
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9MHA2|M9MHA2_9VIRU Replication protein OS=uncultured marine virus OX=186617 PE=4 SV=1
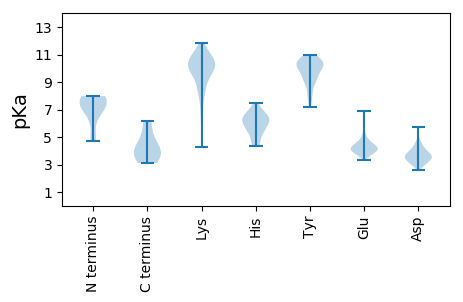
MM1 pKa = 7.98RR2 pKa = 11.84SQHH5 pKa = 5.85QFSQVPRR12 pKa = 11.84ADD14 pKa = 3.13IARR17 pKa = 11.84SSFNLSHH24 pKa = 6.2GVKK27 pKa = 9.11STFDD31 pKa = 3.24VDD33 pKa = 3.96FLVPILVMDD42 pKa = 5.74IIPGDD47 pKa = 3.8TFNVKK52 pKa = 10.24CSFFMRR58 pKa = 11.84LATPLHH64 pKa = 6.26PLMDD68 pKa = 3.62NMYY71 pKa = 10.08VEE73 pKa = 4.53TFFFFVAYY81 pKa = 8.37RR82 pKa = 11.84TIWDD86 pKa = 3.79SFEE89 pKa = 4.34KK90 pKa = 10.64FHH92 pKa = 7.04GAQDD96 pKa = 3.99DD97 pKa = 4.17PGDD100 pKa = 4.66SIDD103 pKa = 3.59FTIPVLSAGIVGTGDD118 pKa = 3.0LSDD121 pKa = 4.09YY122 pKa = 10.47MGLPLALDD130 pKa = 3.6ASVTNDD136 pKa = 2.97VSSLPFRR143 pKa = 11.84AYY145 pKa = 10.48RR146 pKa = 11.84KK147 pKa = 9.04IWNDD151 pKa = 2.61WFRR154 pKa = 11.84DD155 pKa = 3.47EE156 pKa = 5.31NLQDD160 pKa = 3.51TLDD163 pKa = 3.9FNTNNGPDD171 pKa = 3.48NNTNSGMADD180 pKa = 2.96IPLKK184 pKa = 10.01RR185 pKa = 11.84GKK187 pKa = 10.37RR188 pKa = 11.84FDD190 pKa = 4.01YY191 pKa = 8.25FTSSLPAPQKK201 pKa = 10.56GSAVALPLGTRR212 pKa = 11.84ASIATDD218 pKa = 3.42AAAGGLITVKK228 pKa = 10.43ALHH231 pKa = 6.73PALLTQNQLDD241 pKa = 3.8AGGATVDD248 pKa = 4.42LSATTGATGDD258 pKa = 3.95EE259 pKa = 4.44LFADD263 pKa = 4.69LTNATAATINDD274 pKa = 3.0IRR276 pKa = 11.84LAFQTQKK283 pKa = 10.97LIEE286 pKa = 4.58RR287 pKa = 11.84DD288 pKa = 3.39ARR290 pKa = 11.84SGTRR294 pKa = 11.84YY295 pKa = 10.06VEE297 pKa = 3.87TLKK300 pKa = 11.02AHH302 pKa = 6.67WGVTSPDD309 pKa = 3.12HH310 pKa = 6.28RR311 pKa = 11.84LQRR314 pKa = 11.84AEE316 pKa = 3.84FLGGGSSGINITPVAQTSGQTAPVDD341 pKa = 3.64ADD343 pKa = 3.78TMGMLAGFGTASGTHH358 pKa = 5.93GFSKK362 pKa = 10.8SFVEE366 pKa = 4.05HH367 pKa = 6.3GVLIGICNVRR377 pKa = 11.84GDD379 pKa = 3.41ITYY382 pKa = 10.37SQGIDD387 pKa = 3.53RR388 pKa = 11.84MWSKK392 pKa = 9.38STRR395 pKa = 11.84FDD397 pKa = 3.27FFIRR401 pKa = 11.84SSRR404 pKa = 11.84KK405 pKa = 8.43LASRR409 pKa = 11.84RR410 pKa = 11.84YY411 pKa = 9.16SIKK414 pKa = 10.62NMVGRR419 pKa = 11.84EE420 pKa = 4.12CEE422 pKa = 3.62RR423 pKa = 11.84SISVRR428 pKa = 11.84LSRR431 pKa = 11.84KK432 pKa = 6.65VCRR435 pKa = 11.84ISACTIAFMRR445 pKa = 11.84IVV447 pKa = 3.11
MM1 pKa = 7.98RR2 pKa = 11.84SQHH5 pKa = 5.85QFSQVPRR12 pKa = 11.84ADD14 pKa = 3.13IARR17 pKa = 11.84SSFNLSHH24 pKa = 6.2GVKK27 pKa = 9.11STFDD31 pKa = 3.24VDD33 pKa = 3.96FLVPILVMDD42 pKa = 5.74IIPGDD47 pKa = 3.8TFNVKK52 pKa = 10.24CSFFMRR58 pKa = 11.84LATPLHH64 pKa = 6.26PLMDD68 pKa = 3.62NMYY71 pKa = 10.08VEE73 pKa = 4.53TFFFFVAYY81 pKa = 8.37RR82 pKa = 11.84TIWDD86 pKa = 3.79SFEE89 pKa = 4.34KK90 pKa = 10.64FHH92 pKa = 7.04GAQDD96 pKa = 3.99DD97 pKa = 4.17PGDD100 pKa = 4.66SIDD103 pKa = 3.59FTIPVLSAGIVGTGDD118 pKa = 3.0LSDD121 pKa = 4.09YY122 pKa = 10.47MGLPLALDD130 pKa = 3.6ASVTNDD136 pKa = 2.97VSSLPFRR143 pKa = 11.84AYY145 pKa = 10.48RR146 pKa = 11.84KK147 pKa = 9.04IWNDD151 pKa = 2.61WFRR154 pKa = 11.84DD155 pKa = 3.47EE156 pKa = 5.31NLQDD160 pKa = 3.51TLDD163 pKa = 3.9FNTNNGPDD171 pKa = 3.48NNTNSGMADD180 pKa = 2.96IPLKK184 pKa = 10.01RR185 pKa = 11.84GKK187 pKa = 10.37RR188 pKa = 11.84FDD190 pKa = 4.01YY191 pKa = 8.25FTSSLPAPQKK201 pKa = 10.56GSAVALPLGTRR212 pKa = 11.84ASIATDD218 pKa = 3.42AAAGGLITVKK228 pKa = 10.43ALHH231 pKa = 6.73PALLTQNQLDD241 pKa = 3.8AGGATVDD248 pKa = 4.42LSATTGATGDD258 pKa = 3.95EE259 pKa = 4.44LFADD263 pKa = 4.69LTNATAATINDD274 pKa = 3.0IRR276 pKa = 11.84LAFQTQKK283 pKa = 10.97LIEE286 pKa = 4.58RR287 pKa = 11.84DD288 pKa = 3.39ARR290 pKa = 11.84SGTRR294 pKa = 11.84YY295 pKa = 10.06VEE297 pKa = 3.87TLKK300 pKa = 11.02AHH302 pKa = 6.67WGVTSPDD309 pKa = 3.12HH310 pKa = 6.28RR311 pKa = 11.84LQRR314 pKa = 11.84AEE316 pKa = 3.84FLGGGSSGINITPVAQTSGQTAPVDD341 pKa = 3.64ADD343 pKa = 3.78TMGMLAGFGTASGTHH358 pKa = 5.93GFSKK362 pKa = 10.8SFVEE366 pKa = 4.05HH367 pKa = 6.3GVLIGICNVRR377 pKa = 11.84GDD379 pKa = 3.41ITYY382 pKa = 10.37SQGIDD387 pKa = 3.53RR388 pKa = 11.84MWSKK392 pKa = 9.38STRR395 pKa = 11.84FDD397 pKa = 3.27FFIRR401 pKa = 11.84SSRR404 pKa = 11.84KK405 pKa = 8.43LASRR409 pKa = 11.84RR410 pKa = 11.84YY411 pKa = 9.16SIKK414 pKa = 10.62NMVGRR419 pKa = 11.84EE420 pKa = 4.12CEE422 pKa = 3.62RR423 pKa = 11.84SISVRR428 pKa = 11.84LSRR431 pKa = 11.84KK432 pKa = 6.65VCRR435 pKa = 11.84ISACTIAFMRR445 pKa = 11.84IVV447 pKa = 3.11
Molecular weight: 48.9 kDa
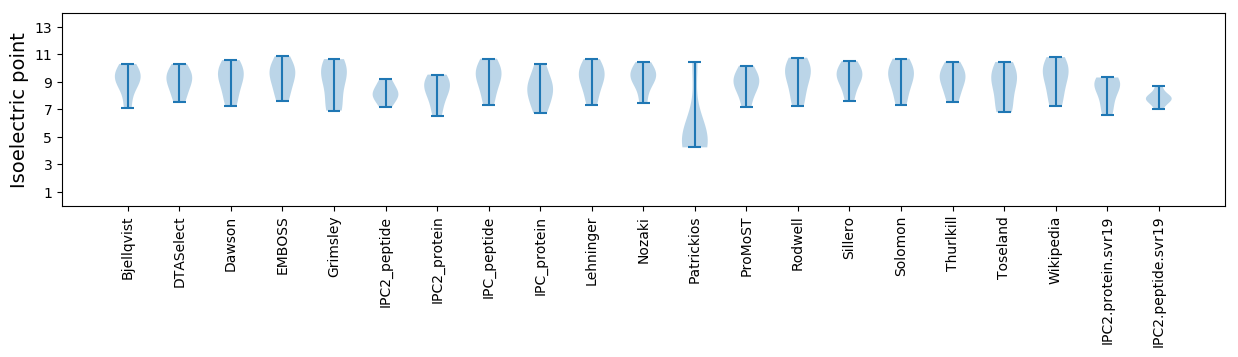
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9M201|M9M201_9VIRU Replication protein (Fragment) OS=uncultured marine virus OX=186617 PE=4 SV=1
DDD2 pKa = 4.72GGSLTWSDDD11 pKa = 3.38ALEEE15 pKa = 4.61DDD17 pKa = 4.87SAALAPNVNDDD28 pKa = 3.15RR29 pKa = 11.84LAFATQHHH37 pKa = 5.4LEEE40 pKa = 4.57DDD42 pKa = 3.38RR43 pKa = 11.84SGTRR47 pKa = 11.84YYY49 pKa = 9.92EEE51 pKa = 4.08LKKK54 pKa = 10.9RR55 pKa = 11.84WGVTSPDDD63 pKa = 3.1RR64 pKa = 11.84LQRR67 pKa = 11.84AEEE70 pKa = 3.84YY71 pKa = 10.06GGGSTRR77 pKa = 11.84INVAPVQQNTASTTPTSPAAQDDD100 pKa = 3.48KK101 pKa = 10.41GNLAGVGTANGTHHH115 pKa = 6.47WSKKK119 pKa = 10.94FVEEE123 pKa = 4.24HH124 pKa = 6.06VIIILGNLRR133 pKa = 11.84GDDD136 pKa = 3.2SYYY139 pKa = 10.53QGVDDD144 pKa = 3.47YYY146 pKa = 9.35SKKK149 pKa = 11.43NTLRR153 pKa = 11.84FRR155 pKa = 11.84ISRR158 pKa = 11.84NGKKK162 pKa = 7.78HH163 pKa = 5.77RR164 pKa = 11.84TSRR167 pKa = 11.84VKKK170 pKa = 10.35RR171 pKa = 11.84NLDDD175 pKa = 3.05HH176 pKa = 6.95HHH178 pKa = 5.49HHH180 pKa = 4.45CH
DDD2 pKa = 4.72GGSLTWSDDD11 pKa = 3.38ALEEE15 pKa = 4.61DDD17 pKa = 4.87SAALAPNVNDDD28 pKa = 3.15RR29 pKa = 11.84LAFATQHHH37 pKa = 5.4LEEE40 pKa = 4.57DDD42 pKa = 3.38RR43 pKa = 11.84SGTRR47 pKa = 11.84YYY49 pKa = 9.92EEE51 pKa = 4.08LKKK54 pKa = 10.9RR55 pKa = 11.84WGVTSPDDD63 pKa = 3.1RR64 pKa = 11.84LQRR67 pKa = 11.84AEEE70 pKa = 3.84YY71 pKa = 10.06GGGSTRR77 pKa = 11.84INVAPVQQNTASTTPTSPAAQDDD100 pKa = 3.48KK101 pKa = 10.41GNLAGVGTANGTHHH115 pKa = 6.47WSKKK119 pKa = 10.94FVEEE123 pKa = 4.24HH124 pKa = 6.06VIIILGNLRR133 pKa = 11.84GDDD136 pKa = 3.2SYYY139 pKa = 10.53QGVDDD144 pKa = 3.47YYY146 pKa = 9.35SKKK149 pKa = 11.43NTLRR153 pKa = 11.84FRR155 pKa = 11.84ISRR158 pKa = 11.84NGKKK162 pKa = 7.78HH163 pKa = 5.77RR164 pKa = 11.84TSRR167 pKa = 11.84VKKK170 pKa = 10.35RR171 pKa = 11.84NLDDD175 pKa = 3.05HH176 pKa = 6.95HHH178 pKa = 5.49HHH180 pKa = 4.45CH
Molecular weight: 20.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1743 |
162 |
447 |
290.5 |
32.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.09 ± 0.468 | 1.721 ± 0.24 |
6.368 ± 0.458 | 3.729 ± 0.564 |
4.246 ± 0.623 | 7.803 ± 0.156 |
3.385 ± 0.477 | 6.024 ± 0.136 |
4.705 ± 0.661 | 7.057 ± 0.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.328 ± 0.434 | 4.705 ± 0.256 |
4.59 ± 0.298 | 2.754 ± 0.153 |
8.032 ± 0.503 | 5.565 ± 1.012 |
6.942 ± 0.467 | 5.106 ± 0.381 |
2.811 ± 0.523 | 2.983 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
