
Potato yellow dwarf nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
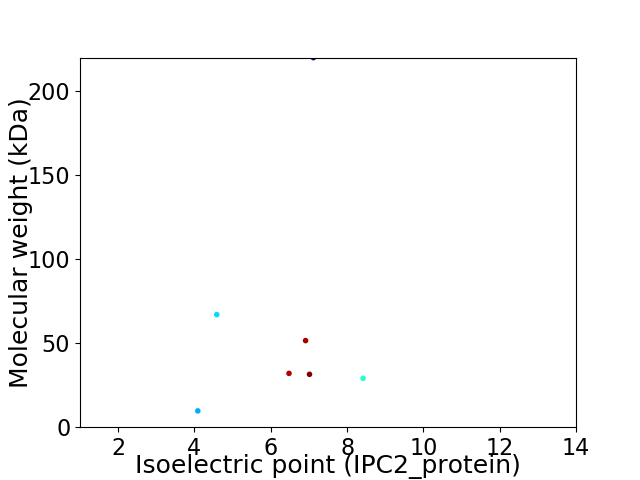
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5L202|D5L202_9RHAB Putative movement protein OS=Potato yellow dwarf nucleorhabdovirus OX=195060 GN=Y PE=4 SV=1
MM1 pKa = 7.61EE2 pKa = 6.02SDD4 pKa = 3.62SATPGQGKK12 pKa = 8.23PAATDD17 pKa = 3.39HH18 pKa = 6.66KK19 pKa = 9.99PPEE22 pKa = 4.42PTPTTAADD30 pKa = 3.48PTKK33 pKa = 10.34QQEE36 pKa = 4.68TKK38 pKa = 10.38EE39 pKa = 4.46SPDD42 pKa = 3.12QHH44 pKa = 7.27GKK46 pKa = 9.83EE47 pKa = 3.9EE48 pKa = 4.16HH49 pKa = 6.02TEE51 pKa = 4.0EE52 pKa = 4.61EE53 pKa = 4.13KK54 pKa = 11.13DD55 pKa = 3.63STIEE59 pKa = 3.62WVYY62 pKa = 10.98PDD64 pKa = 4.24WDD66 pKa = 4.82DD67 pKa = 4.09NDD69 pKa = 3.98SDD71 pKa = 5.03IYY73 pKa = 11.53DD74 pKa = 4.19LLYY77 pKa = 10.75EE78 pKa = 4.69CNHH81 pKa = 6.28GEE83 pKa = 3.95WGNN86 pKa = 3.42
MM1 pKa = 7.61EE2 pKa = 6.02SDD4 pKa = 3.62SATPGQGKK12 pKa = 8.23PAATDD17 pKa = 3.39HH18 pKa = 6.66KK19 pKa = 9.99PPEE22 pKa = 4.42PTPTTAADD30 pKa = 3.48PTKK33 pKa = 10.34QQEE36 pKa = 4.68TKK38 pKa = 10.38EE39 pKa = 4.46SPDD42 pKa = 3.12QHH44 pKa = 7.27GKK46 pKa = 9.83EE47 pKa = 3.9EE48 pKa = 4.16HH49 pKa = 6.02TEE51 pKa = 4.0EE52 pKa = 4.61EE53 pKa = 4.13KK54 pKa = 11.13DD55 pKa = 3.63STIEE59 pKa = 3.62WVYY62 pKa = 10.98PDD64 pKa = 4.24WDD66 pKa = 4.82DD67 pKa = 4.09NDD69 pKa = 3.98SDD71 pKa = 5.03IYY73 pKa = 11.53DD74 pKa = 4.19LLYY77 pKa = 10.75EE78 pKa = 4.69CNHH81 pKa = 6.28GEE83 pKa = 3.95WGNN86 pKa = 3.42
Molecular weight: 9.7 kDa
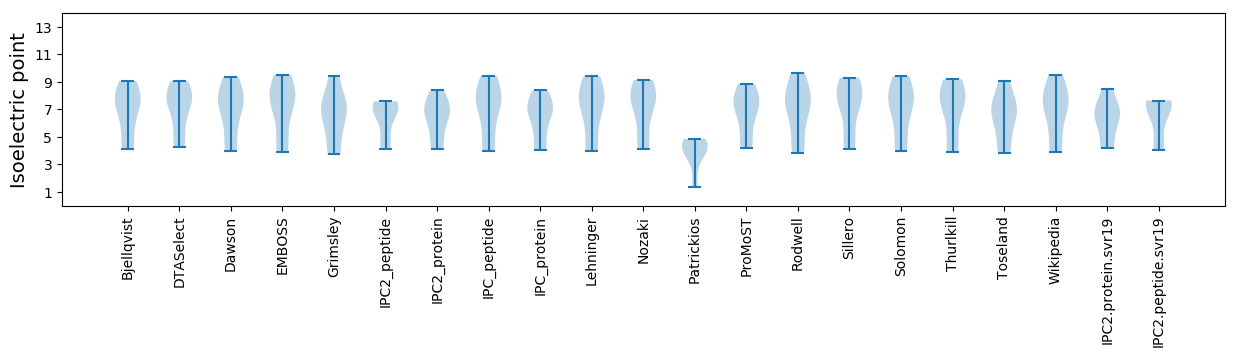
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5L204|D5L204_9RHAB Glycoprotein OS=Potato yellow dwarf nucleorhabdovirus OX=195060 GN=G PE=4 SV=1
MM1 pKa = 8.01ISMIGSRR8 pKa = 11.84RR9 pKa = 11.84PIRR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 3.44VYY16 pKa = 9.24LASKK20 pKa = 9.73EE21 pKa = 4.42GEE23 pKa = 3.99VTLQKK28 pKa = 10.61EE29 pKa = 4.1KK30 pKa = 10.47TGILISLNYY39 pKa = 8.74IVKK42 pKa = 9.48IHH44 pKa = 6.96LYY46 pKa = 10.6DD47 pKa = 3.51LVLRR51 pKa = 11.84DD52 pKa = 3.93EE53 pKa = 5.15IEE55 pKa = 4.19NQGISYY61 pKa = 10.12PDD63 pKa = 3.24LFKK66 pKa = 11.23YY67 pKa = 11.02VEE69 pKa = 4.3DD70 pKa = 4.7QIGLPPNNPEE80 pKa = 4.36AIIRR84 pKa = 11.84SPDD87 pKa = 3.29MTNSQVKK94 pKa = 9.17EE95 pKa = 4.2LIMLAKK101 pKa = 9.65LHH103 pKa = 6.64AMHH106 pKa = 6.88TEE108 pKa = 3.8EE109 pKa = 5.43KK110 pKa = 9.53ITVFTQKK117 pKa = 9.56TDD119 pKa = 3.81FSEE122 pKa = 4.28KK123 pKa = 10.3TPTLYY128 pKa = 11.01LKK130 pKa = 10.66VGEE133 pKa = 4.49SILRR137 pKa = 11.84NDD139 pKa = 4.0HH140 pKa = 6.88DD141 pKa = 3.91PHH143 pKa = 7.42VVEE146 pKa = 5.14VIYY149 pKa = 10.6RR150 pKa = 11.84DD151 pKa = 3.73SDD153 pKa = 3.83PLPKK157 pKa = 10.4GEE159 pKa = 4.3YY160 pKa = 8.13QLNMTRR166 pKa = 11.84QYY168 pKa = 11.22KK169 pKa = 10.5GDD171 pKa = 3.58SDD173 pKa = 4.08QIKK176 pKa = 10.71CEE178 pKa = 3.53ITFAMFVRR186 pKa = 11.84TPPRR190 pKa = 11.84NNNHH194 pKa = 4.21VSARR198 pKa = 11.84LNLMVLLSKK207 pKa = 10.91YY208 pKa = 10.87SNEE211 pKa = 3.91LKK213 pKa = 9.69KK214 pKa = 9.28TVSDD218 pKa = 3.84PLKK221 pKa = 10.7NLLKK225 pKa = 10.37RR226 pKa = 11.84KK227 pKa = 9.85SDD229 pKa = 3.29TTIPTKK235 pKa = 10.51TDD237 pKa = 2.79GSKK240 pKa = 10.49RR241 pKa = 11.84AFLPSMLKK249 pKa = 9.32MLSPP253 pKa = 4.61
MM1 pKa = 8.01ISMIGSRR8 pKa = 11.84RR9 pKa = 11.84PIRR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 3.44VYY16 pKa = 9.24LASKK20 pKa = 9.73EE21 pKa = 4.42GEE23 pKa = 3.99VTLQKK28 pKa = 10.61EE29 pKa = 4.1KK30 pKa = 10.47TGILISLNYY39 pKa = 8.74IVKK42 pKa = 9.48IHH44 pKa = 6.96LYY46 pKa = 10.6DD47 pKa = 3.51LVLRR51 pKa = 11.84DD52 pKa = 3.93EE53 pKa = 5.15IEE55 pKa = 4.19NQGISYY61 pKa = 10.12PDD63 pKa = 3.24LFKK66 pKa = 11.23YY67 pKa = 11.02VEE69 pKa = 4.3DD70 pKa = 4.7QIGLPPNNPEE80 pKa = 4.36AIIRR84 pKa = 11.84SPDD87 pKa = 3.29MTNSQVKK94 pKa = 9.17EE95 pKa = 4.2LIMLAKK101 pKa = 9.65LHH103 pKa = 6.64AMHH106 pKa = 6.88TEE108 pKa = 3.8EE109 pKa = 5.43KK110 pKa = 9.53ITVFTQKK117 pKa = 9.56TDD119 pKa = 3.81FSEE122 pKa = 4.28KK123 pKa = 10.3TPTLYY128 pKa = 11.01LKK130 pKa = 10.66VGEE133 pKa = 4.49SILRR137 pKa = 11.84NDD139 pKa = 4.0HH140 pKa = 6.88DD141 pKa = 3.91PHH143 pKa = 7.42VVEE146 pKa = 5.14VIYY149 pKa = 10.6RR150 pKa = 11.84DD151 pKa = 3.73SDD153 pKa = 3.83PLPKK157 pKa = 10.4GEE159 pKa = 4.3YY160 pKa = 8.13QLNMTRR166 pKa = 11.84QYY168 pKa = 11.22KK169 pKa = 10.5GDD171 pKa = 3.58SDD173 pKa = 4.08QIKK176 pKa = 10.71CEE178 pKa = 3.53ITFAMFVRR186 pKa = 11.84TPPRR190 pKa = 11.84NNNHH194 pKa = 4.21VSARR198 pKa = 11.84LNLMVLLSKK207 pKa = 10.91YY208 pKa = 10.87SNEE211 pKa = 3.91LKK213 pKa = 9.69KK214 pKa = 9.28TVSDD218 pKa = 3.84PLKK221 pKa = 10.7NLLKK225 pKa = 10.37RR226 pKa = 11.84KK227 pKa = 9.85SDD229 pKa = 3.29TTIPTKK235 pKa = 10.51TDD237 pKa = 2.79GSKK240 pKa = 10.49RR241 pKa = 11.84AFLPSMLKK249 pKa = 9.32MLSPP253 pKa = 4.61
Molecular weight: 29.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3914 |
86 |
1931 |
559.1 |
62.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.212 ± 1.103 | 1.533 ± 0.287 |
5.825 ± 0.554 | 5.544 ± 0.799 |
3.602 ± 0.301 | 5.8 ± 0.37 |
1.993 ± 0.324 | 7.154 ± 0.503 |
5.442 ± 0.716 | 9.274 ± 0.786 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.577 ± 0.589 | 5.263 ± 0.368 |
5.11 ± 0.636 | 3.168 ± 0.425 |
5.314 ± 0.679 | 8.278 ± 0.976 |
7.052 ± 0.668 | 5.953 ± 0.576 |
1.252 ± 0.312 | 3.654 ± 0.185 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
