
Dragonfly associated cyclovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins
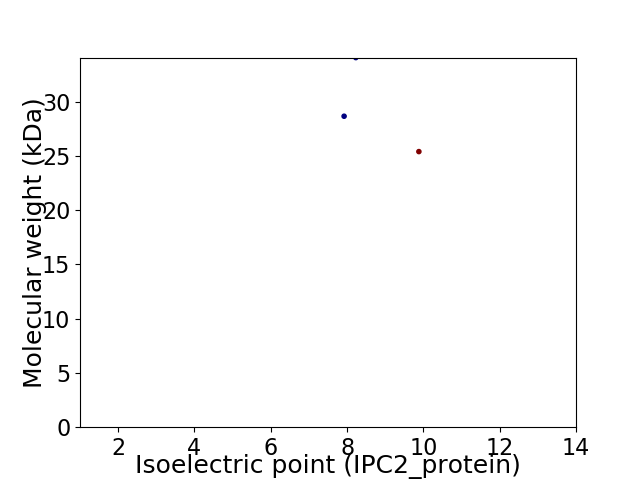
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
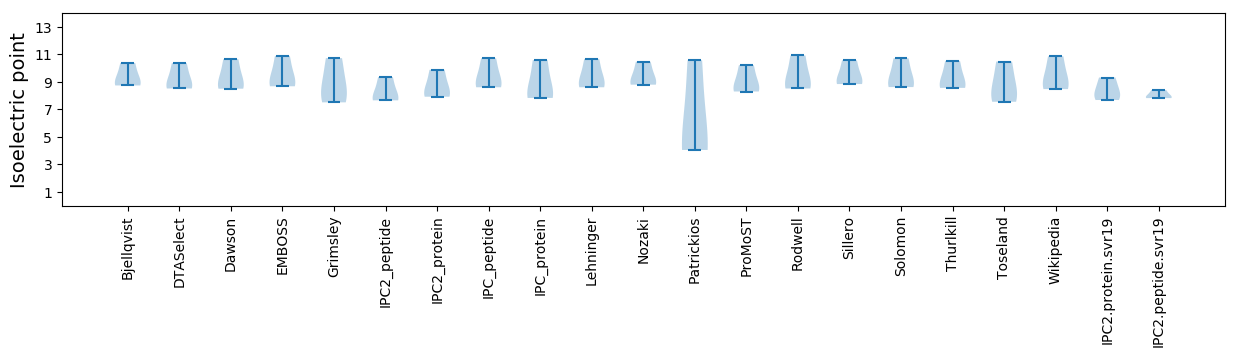
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A1K7|K0A1K7_9CIRC Spliced replication-associated protein OS=Dragonfly associated cyclovirus 3 OX=1234881 PE=4 SV=1
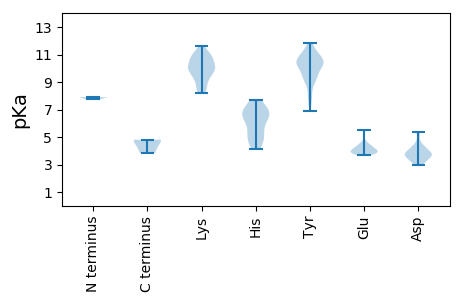
MM1 pKa = 7.9PSPRR5 pKa = 11.84DD6 pKa = 3.16LKK8 pKa = 10.82GAVRR12 pKa = 11.84LGIYY16 pKa = 8.15VLRR19 pKa = 11.84CLYY22 pKa = 10.42NGTMSARR29 pKa = 11.84QRR31 pKa = 11.84LKK33 pKa = 10.78RR34 pKa = 11.84IVFTLNNYY42 pKa = 10.06CEE44 pKa = 4.55DD45 pKa = 3.86DD46 pKa = 3.88FVRR49 pKa = 11.84IQSYY53 pKa = 9.01VEE55 pKa = 3.95IYY57 pKa = 9.68QYY59 pKa = 11.1CVVGRR64 pKa = 11.84EE65 pKa = 4.18TAPEE69 pKa = 4.04TGTPHH74 pKa = 6.13LQGFINFKK82 pKa = 9.91SKK84 pKa = 11.2RR85 pKa = 11.84EE86 pKa = 3.75FGTIKK91 pKa = 10.07TIVGDD96 pKa = 3.69RR97 pKa = 11.84AHH99 pKa = 7.14IEE101 pKa = 3.85PAKK104 pKa = 10.88GSDD107 pKa = 3.6VQNKK111 pKa = 8.23EE112 pKa = 3.92YY113 pKa = 9.25CTKK116 pKa = 10.92GGDD119 pKa = 3.41YY120 pKa = 9.68WEE122 pKa = 5.54CGTPSGPGYY131 pKa = 10.44RR132 pKa = 11.84SDD134 pKa = 4.17LADD137 pKa = 3.11VVQTVKK143 pKa = 10.32GAKK146 pKa = 9.34RR147 pKa = 11.84LRR149 pKa = 11.84EE150 pKa = 3.89VVEE153 pKa = 3.76RR154 pKa = 11.84HH155 pKa = 4.82PCEE158 pKa = 3.95FIKK161 pKa = 9.71YY162 pKa = 9.32HH163 pKa = 6.67RR164 pKa = 11.84GIEE167 pKa = 4.14KK168 pKa = 10.41LFGMLSEE175 pKa = 4.2RR176 pKa = 11.84EE177 pKa = 3.79KK178 pKa = 11.16RR179 pKa = 11.84NWKK182 pKa = 8.47TEE184 pKa = 4.04TIVYY188 pKa = 10.28YY189 pKa = 10.74GDD191 pKa = 3.83PGAGKK196 pKa = 9.66SRR198 pKa = 11.84KK199 pKa = 9.0AAEE202 pKa = 3.91VGAAAEE208 pKa = 4.23GGVYY212 pKa = 10.49YY213 pKa = 9.22KK214 pKa = 10.39TRR216 pKa = 11.84GPWWDD221 pKa = 3.66GYY223 pKa = 11.25NGEE226 pKa = 4.39TTVIVDD232 pKa = 5.39DD233 pKa = 4.36YY234 pKa = 10.71YY235 pKa = 11.83GWLAYY240 pKa = 10.53DD241 pKa = 4.01EE242 pKa = 4.4VLKK245 pKa = 9.99ITDD248 pKa = 3.59RR249 pKa = 11.84YY250 pKa = 10.33LL251 pKa = 4.8
MM1 pKa = 7.9PSPRR5 pKa = 11.84DD6 pKa = 3.16LKK8 pKa = 10.82GAVRR12 pKa = 11.84LGIYY16 pKa = 8.15VLRR19 pKa = 11.84CLYY22 pKa = 10.42NGTMSARR29 pKa = 11.84QRR31 pKa = 11.84LKK33 pKa = 10.78RR34 pKa = 11.84IVFTLNNYY42 pKa = 10.06CEE44 pKa = 4.55DD45 pKa = 3.86DD46 pKa = 3.88FVRR49 pKa = 11.84IQSYY53 pKa = 9.01VEE55 pKa = 3.95IYY57 pKa = 9.68QYY59 pKa = 11.1CVVGRR64 pKa = 11.84EE65 pKa = 4.18TAPEE69 pKa = 4.04TGTPHH74 pKa = 6.13LQGFINFKK82 pKa = 9.91SKK84 pKa = 11.2RR85 pKa = 11.84EE86 pKa = 3.75FGTIKK91 pKa = 10.07TIVGDD96 pKa = 3.69RR97 pKa = 11.84AHH99 pKa = 7.14IEE101 pKa = 3.85PAKK104 pKa = 10.88GSDD107 pKa = 3.6VQNKK111 pKa = 8.23EE112 pKa = 3.92YY113 pKa = 9.25CTKK116 pKa = 10.92GGDD119 pKa = 3.41YY120 pKa = 9.68WEE122 pKa = 5.54CGTPSGPGYY131 pKa = 10.44RR132 pKa = 11.84SDD134 pKa = 4.17LADD137 pKa = 3.11VVQTVKK143 pKa = 10.32GAKK146 pKa = 9.34RR147 pKa = 11.84LRR149 pKa = 11.84EE150 pKa = 3.89VVEE153 pKa = 3.76RR154 pKa = 11.84HH155 pKa = 4.82PCEE158 pKa = 3.95FIKK161 pKa = 9.71YY162 pKa = 9.32HH163 pKa = 6.67RR164 pKa = 11.84GIEE167 pKa = 4.14KK168 pKa = 10.41LFGMLSEE175 pKa = 4.2RR176 pKa = 11.84EE177 pKa = 3.79KK178 pKa = 11.16RR179 pKa = 11.84NWKK182 pKa = 8.47TEE184 pKa = 4.04TIVYY188 pKa = 10.28YY189 pKa = 10.74GDD191 pKa = 3.83PGAGKK196 pKa = 9.66SRR198 pKa = 11.84KK199 pKa = 9.0AAEE202 pKa = 3.91VGAAAEE208 pKa = 4.23GGVYY212 pKa = 10.49YY213 pKa = 9.22KK214 pKa = 10.39TRR216 pKa = 11.84GPWWDD221 pKa = 3.66GYY223 pKa = 11.25NGEE226 pKa = 4.39TTVIVDD232 pKa = 5.39DD233 pKa = 4.36YY234 pKa = 10.71YY235 pKa = 11.83GWLAYY240 pKa = 10.53DD241 pKa = 4.01EE242 pKa = 4.4VLKK245 pKa = 9.99ITDD248 pKa = 3.59RR249 pKa = 11.84YY250 pKa = 10.33LL251 pKa = 4.8
Molecular weight: 28.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A154|K0A154_9CIRC Replication-associated protein OS=Dragonfly associated cyclovirus 3 OX=1234881 PE=4 SV=1
MM1 pKa = 7.74AYY3 pKa = 10.44LRR5 pKa = 11.84ARR7 pKa = 11.84YY8 pKa = 6.89TNRR11 pKa = 11.84RR12 pKa = 11.84FSRR15 pKa = 11.84RR16 pKa = 11.84GLRR19 pKa = 11.84QVQRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84LIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84KK35 pKa = 9.96SNLSCKK41 pKa = 9.1FTTNLVLKK49 pKa = 8.44QTTGIQYY56 pKa = 11.14AHH58 pKa = 7.68IAPALEE64 pKa = 4.82DD65 pKa = 3.67FAEE68 pKa = 4.42TTNLITNFEE77 pKa = 4.39YY78 pKa = 11.07YY79 pKa = 10.45KK80 pKa = 10.06IRR82 pKa = 11.84KK83 pKa = 8.51VRR85 pKa = 11.84ITVTPRR91 pKa = 11.84NNVAYY96 pKa = 10.1QGQTVPDD103 pKa = 3.75YY104 pKa = 11.24VIAPFHH110 pKa = 6.76HH111 pKa = 6.77PVDD114 pKa = 4.46DD115 pKa = 4.33KK116 pKa = 11.64SISVDD121 pKa = 3.33SLLTLDD127 pKa = 3.23RR128 pKa = 11.84HH129 pKa = 5.37KK130 pKa = 10.66KK131 pKa = 9.38YY132 pKa = 10.59RR133 pKa = 11.84GTQRR137 pKa = 11.84GHH139 pKa = 4.75MTFKK143 pKa = 10.31PAVIGLASTSLDD155 pKa = 3.7DD156 pKa = 4.94SSGTYY161 pKa = 9.1ATMRR165 pKa = 11.84WSPKK169 pKa = 10.12ILITNNTTPQQVKK182 pKa = 9.72HH183 pKa = 5.4FCGLLAFAPQGSNAQEE199 pKa = 3.79YY200 pKa = 10.07EE201 pKa = 3.98ITLDD205 pKa = 4.2AYY207 pKa = 10.17CTFYY211 pKa = 10.57NQKK214 pKa = 9.89INKK217 pKa = 8.94LLL219 pKa = 3.84
MM1 pKa = 7.74AYY3 pKa = 10.44LRR5 pKa = 11.84ARR7 pKa = 11.84YY8 pKa = 6.89TNRR11 pKa = 11.84RR12 pKa = 11.84FSRR15 pKa = 11.84RR16 pKa = 11.84GLRR19 pKa = 11.84QVQRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84LIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84KK35 pKa = 9.96SNLSCKK41 pKa = 9.1FTTNLVLKK49 pKa = 8.44QTTGIQYY56 pKa = 11.14AHH58 pKa = 7.68IAPALEE64 pKa = 4.82DD65 pKa = 3.67FAEE68 pKa = 4.42TTNLITNFEE77 pKa = 4.39YY78 pKa = 11.07YY79 pKa = 10.45KK80 pKa = 10.06IRR82 pKa = 11.84KK83 pKa = 8.51VRR85 pKa = 11.84ITVTPRR91 pKa = 11.84NNVAYY96 pKa = 10.1QGQTVPDD103 pKa = 3.75YY104 pKa = 11.24VIAPFHH110 pKa = 6.76HH111 pKa = 6.77PVDD114 pKa = 4.46DD115 pKa = 4.33KK116 pKa = 11.64SISVDD121 pKa = 3.33SLLTLDD127 pKa = 3.23RR128 pKa = 11.84HH129 pKa = 5.37KK130 pKa = 10.66KK131 pKa = 9.38YY132 pKa = 10.59RR133 pKa = 11.84GTQRR137 pKa = 11.84GHH139 pKa = 4.75MTFKK143 pKa = 10.31PAVIGLASTSLDD155 pKa = 3.7DD156 pKa = 4.94SSGTYY161 pKa = 9.1ATMRR165 pKa = 11.84WSPKK169 pKa = 10.12ILITNNTTPQQVKK182 pKa = 9.72HH183 pKa = 5.4FCGLLAFAPQGSNAQEE199 pKa = 3.79YY200 pKa = 10.07EE201 pKa = 3.98ITLDD205 pKa = 4.2AYY207 pKa = 10.17CTFYY211 pKa = 10.57NQKK214 pKa = 9.89INKK217 pKa = 8.94LLL219 pKa = 3.84
Molecular weight: 25.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
770 |
219 |
300 |
256.7 |
29.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.714 ± 0.44 | 1.948 ± 0.229 |
4.935 ± 0.33 | 5.714 ± 1.265 |
3.377 ± 0.296 | 9.091 ± 1.823 |
2.078 ± 0.259 | 5.195 ± 0.3 |
6.494 ± 0.285 | 7.013 ± 0.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.169 ± 0.088 | 3.636 ± 0.672 |
4.286 ± 0.132 | 3.117 ± 0.868 |
8.571 ± 0.709 | 4.416 ± 0.425 |
7.662 ± 0.878 | 7.143 ± 0.773 |
1.688 ± 0.458 | 6.753 ± 0.482 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
