
Sewage-associated circular DNA virus-37
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
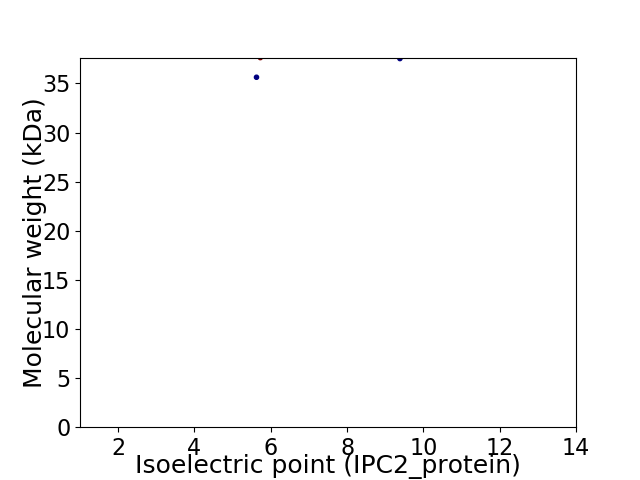
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
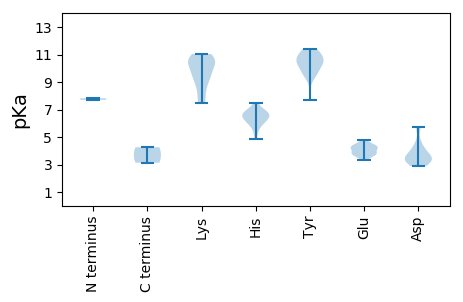
Protein with the lowest isoelectric point:
>tr|A0A0B4UI36|A0A0B4UI36_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-37 OX=1592104 PE=4 SV=1
MM1 pKa = 7.82PFRR4 pKa = 11.84VSAKK8 pKa = 10.28SFIITYY14 pKa = 7.68PRR16 pKa = 11.84CDD18 pKa = 3.1VDD20 pKa = 5.69KK21 pKa = 11.05EE22 pKa = 4.77VILQWFQDD30 pKa = 3.18HH31 pKa = 6.86FMLLGCRR38 pKa = 11.84VARR41 pKa = 11.84EE42 pKa = 3.84LHH44 pKa = 6.72ADD46 pKa = 4.03GTPHH50 pKa = 5.84IHH52 pKa = 6.11VVFHH56 pKa = 6.94IANTFQTRR64 pKa = 11.84NARR67 pKa = 11.84YY68 pKa = 9.2FDD70 pKa = 4.1IGDD73 pKa = 3.5HH74 pKa = 6.32HH75 pKa = 7.48PNVQSTRR82 pKa = 11.84GLPASYY88 pKa = 10.4AYY90 pKa = 9.75VAKK93 pKa = 10.81DD94 pKa = 2.94GDD96 pKa = 4.34YY97 pKa = 10.68IDD99 pKa = 5.7FGTIPATRR107 pKa = 11.84PHH109 pKa = 6.75GSWADD114 pKa = 3.28IANATTKK121 pKa = 10.72EE122 pKa = 3.73EE123 pKa = 3.79ALALVKK129 pKa = 9.21EE130 pKa = 4.51TSPRR134 pKa = 11.84DD135 pKa = 3.3YY136 pKa = 11.26VLNYY140 pKa = 10.65DD141 pKa = 4.71KK142 pKa = 10.64IISFCEE148 pKa = 3.51SHH150 pKa = 6.3FANRR154 pKa = 11.84TEE156 pKa = 4.2AYY158 pKa = 10.2VPIYY162 pKa = 10.71QNFNGLPQQLTTWCDD177 pKa = 3.01QRR179 pKa = 11.84LEE181 pKa = 4.02VSEE184 pKa = 4.26RR185 pKa = 11.84HH186 pKa = 5.76SPPPAAGLPPQPPTEE201 pKa = 4.1WGEE204 pKa = 3.78LLYY207 pKa = 10.34IVTNQSLGHH216 pKa = 5.97HH217 pKa = 6.45MYY219 pKa = 10.45FNAMFDD225 pKa = 3.41LGEE228 pKa = 4.2WDD230 pKa = 5.63DD231 pKa = 3.91GAQYY235 pKa = 11.16AIFDD239 pKa = 4.36DD240 pKa = 3.71WADD243 pKa = 2.99WSKK246 pKa = 11.03FYY248 pKa = 10.66AYY250 pKa = 9.67KK251 pKa = 10.48QFLGAQRR258 pKa = 11.84QFTITDD264 pKa = 3.49KK265 pKa = 10.76YY266 pKa = 9.4RR267 pKa = 11.84RR268 pKa = 11.84KK269 pKa = 9.33RR270 pKa = 11.84TVRR273 pKa = 11.84WGKK276 pKa = 8.56PAIVISNLRR285 pKa = 11.84PAWEE289 pKa = 4.44DD290 pKa = 3.5EE291 pKa = 3.83DD292 pKa = 4.61WITVNCFQCHH302 pKa = 5.39LLGALFF308 pKa = 4.26
MM1 pKa = 7.82PFRR4 pKa = 11.84VSAKK8 pKa = 10.28SFIITYY14 pKa = 7.68PRR16 pKa = 11.84CDD18 pKa = 3.1VDD20 pKa = 5.69KK21 pKa = 11.05EE22 pKa = 4.77VILQWFQDD30 pKa = 3.18HH31 pKa = 6.86FMLLGCRR38 pKa = 11.84VARR41 pKa = 11.84EE42 pKa = 3.84LHH44 pKa = 6.72ADD46 pKa = 4.03GTPHH50 pKa = 5.84IHH52 pKa = 6.11VVFHH56 pKa = 6.94IANTFQTRR64 pKa = 11.84NARR67 pKa = 11.84YY68 pKa = 9.2FDD70 pKa = 4.1IGDD73 pKa = 3.5HH74 pKa = 6.32HH75 pKa = 7.48PNVQSTRR82 pKa = 11.84GLPASYY88 pKa = 10.4AYY90 pKa = 9.75VAKK93 pKa = 10.81DD94 pKa = 2.94GDD96 pKa = 4.34YY97 pKa = 10.68IDD99 pKa = 5.7FGTIPATRR107 pKa = 11.84PHH109 pKa = 6.75GSWADD114 pKa = 3.28IANATTKK121 pKa = 10.72EE122 pKa = 3.73EE123 pKa = 3.79ALALVKK129 pKa = 9.21EE130 pKa = 4.51TSPRR134 pKa = 11.84DD135 pKa = 3.3YY136 pKa = 11.26VLNYY140 pKa = 10.65DD141 pKa = 4.71KK142 pKa = 10.64IISFCEE148 pKa = 3.51SHH150 pKa = 6.3FANRR154 pKa = 11.84TEE156 pKa = 4.2AYY158 pKa = 10.2VPIYY162 pKa = 10.71QNFNGLPQQLTTWCDD177 pKa = 3.01QRR179 pKa = 11.84LEE181 pKa = 4.02VSEE184 pKa = 4.26RR185 pKa = 11.84HH186 pKa = 5.76SPPPAAGLPPQPPTEE201 pKa = 4.1WGEE204 pKa = 3.78LLYY207 pKa = 10.34IVTNQSLGHH216 pKa = 5.97HH217 pKa = 6.45MYY219 pKa = 10.45FNAMFDD225 pKa = 3.41LGEE228 pKa = 4.2WDD230 pKa = 5.63DD231 pKa = 3.91GAQYY235 pKa = 11.16AIFDD239 pKa = 4.36DD240 pKa = 3.71WADD243 pKa = 2.99WSKK246 pKa = 11.03FYY248 pKa = 10.66AYY250 pKa = 9.67KK251 pKa = 10.48QFLGAQRR258 pKa = 11.84QFTITDD264 pKa = 3.49KK265 pKa = 10.76YY266 pKa = 9.4RR267 pKa = 11.84RR268 pKa = 11.84KK269 pKa = 9.33RR270 pKa = 11.84TVRR273 pKa = 11.84WGKK276 pKa = 8.56PAIVISNLRR285 pKa = 11.84PAWEE289 pKa = 4.44DD290 pKa = 3.5EE291 pKa = 3.83DD292 pKa = 4.61WITVNCFQCHH302 pKa = 5.39LLGALFF308 pKa = 4.26
Molecular weight: 35.63 kDa
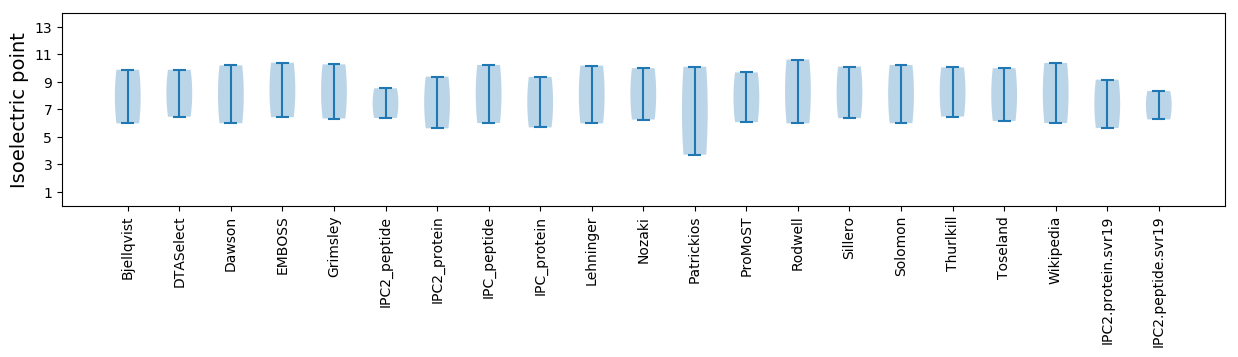
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI36|A0A0B4UI36_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-37 OX=1592104 PE=4 SV=1
MM1 pKa = 7.7PHH3 pKa = 6.6RR4 pKa = 11.84NWRR7 pKa = 11.84KK8 pKa = 7.49RR9 pKa = 11.84THH11 pKa = 6.96DD12 pKa = 3.47ALHH15 pKa = 6.3RR16 pKa = 11.84VGSHH20 pKa = 4.85VLRR23 pKa = 11.84NGKK26 pKa = 7.9TYY28 pKa = 11.0GDD30 pKa = 3.71VYY32 pKa = 11.1SAYY35 pKa = 9.85KK36 pKa = 9.95KK37 pKa = 10.77LKK39 pKa = 7.52THH41 pKa = 6.75HH42 pKa = 6.46SSEE45 pKa = 4.38RR46 pKa = 11.84SIPMAPKK53 pKa = 10.36DD54 pKa = 3.45ALTNQFNAQTIFRR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.55KK70 pKa = 7.64NRR72 pKa = 11.84RR73 pKa = 11.84WRR75 pKa = 11.84KK76 pKa = 8.46QLSFAQKK83 pKa = 10.06VLRR86 pKa = 11.84ANQIFSSEE94 pKa = 3.67KK95 pKa = 8.82TYY97 pKa = 11.42QLMRR101 pKa = 11.84TWASAAAAGVQGFNGISLYY120 pKa = 11.4ANGSGDD126 pKa = 4.17ADD128 pKa = 3.63VNDD131 pKa = 3.8IFQAYY136 pKa = 10.31DD137 pKa = 3.11NVTSRR142 pKa = 11.84EE143 pKa = 3.35WSINMKK149 pKa = 9.81SAHH152 pKa = 6.19VDD154 pKa = 3.52YY155 pKa = 11.1LCTNVGSTTLVLKK168 pKa = 10.71VYY170 pKa = 9.65TIVPKK175 pKa = 10.49KK176 pKa = 9.86DD177 pKa = 3.18APVAAGNNLGNAWIASLDD195 pKa = 3.61MSVAMPGSTVGLDD208 pKa = 3.15QIVANPVGTPARR220 pKa = 11.84PGATPFDD227 pKa = 3.77SSVFCSQYY235 pKa = 10.6TVIKK239 pKa = 9.05CVEE242 pKa = 4.41YY243 pKa = 10.24ILPPGQVASFNDD255 pKa = 2.89SMMRR259 pKa = 11.84RR260 pKa = 11.84GKK262 pKa = 10.73LDD264 pKa = 3.26TAEE267 pKa = 4.28FNVGVATTKK276 pKa = 10.44TNWAVKK282 pKa = 9.54GWTKK286 pKa = 10.67SHH288 pKa = 6.58LFVHH292 pKa = 6.6HH293 pKa = 7.09GIATSGAVSSLATYY307 pKa = 10.33YY308 pKa = 9.68PASSLGVIANKK319 pKa = 9.21TYY321 pKa = 10.88KK322 pKa = 9.74FTVNEE327 pKa = 3.71QRR329 pKa = 11.84NQSEE333 pKa = 4.18IQVASTVTT341 pKa = 3.13
MM1 pKa = 7.7PHH3 pKa = 6.6RR4 pKa = 11.84NWRR7 pKa = 11.84KK8 pKa = 7.49RR9 pKa = 11.84THH11 pKa = 6.96DD12 pKa = 3.47ALHH15 pKa = 6.3RR16 pKa = 11.84VGSHH20 pKa = 4.85VLRR23 pKa = 11.84NGKK26 pKa = 7.9TYY28 pKa = 11.0GDD30 pKa = 3.71VYY32 pKa = 11.1SAYY35 pKa = 9.85KK36 pKa = 9.95KK37 pKa = 10.77LKK39 pKa = 7.52THH41 pKa = 6.75HH42 pKa = 6.46SSEE45 pKa = 4.38RR46 pKa = 11.84SIPMAPKK53 pKa = 10.36DD54 pKa = 3.45ALTNQFNAQTIFRR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.55KK70 pKa = 7.64NRR72 pKa = 11.84RR73 pKa = 11.84WRR75 pKa = 11.84KK76 pKa = 8.46QLSFAQKK83 pKa = 10.06VLRR86 pKa = 11.84ANQIFSSEE94 pKa = 3.67KK95 pKa = 8.82TYY97 pKa = 11.42QLMRR101 pKa = 11.84TWASAAAAGVQGFNGISLYY120 pKa = 11.4ANGSGDD126 pKa = 4.17ADD128 pKa = 3.63VNDD131 pKa = 3.8IFQAYY136 pKa = 10.31DD137 pKa = 3.11NVTSRR142 pKa = 11.84EE143 pKa = 3.35WSINMKK149 pKa = 9.81SAHH152 pKa = 6.19VDD154 pKa = 3.52YY155 pKa = 11.1LCTNVGSTTLVLKK168 pKa = 10.71VYY170 pKa = 9.65TIVPKK175 pKa = 10.49KK176 pKa = 9.86DD177 pKa = 3.18APVAAGNNLGNAWIASLDD195 pKa = 3.61MSVAMPGSTVGLDD208 pKa = 3.15QIVANPVGTPARR220 pKa = 11.84PGATPFDD227 pKa = 3.77SSVFCSQYY235 pKa = 10.6TVIKK239 pKa = 9.05CVEE242 pKa = 4.41YY243 pKa = 10.24ILPPGQVASFNDD255 pKa = 2.89SMMRR259 pKa = 11.84RR260 pKa = 11.84GKK262 pKa = 10.73LDD264 pKa = 3.26TAEE267 pKa = 4.28FNVGVATTKK276 pKa = 10.44TNWAVKK282 pKa = 9.54GWTKK286 pKa = 10.67SHH288 pKa = 6.58LFVHH292 pKa = 6.6HH293 pKa = 7.09GIATSGAVSSLATYY307 pKa = 10.33YY308 pKa = 9.68PASSLGVIANKK319 pKa = 9.21TYY321 pKa = 10.88KK322 pKa = 9.74FTVNEE327 pKa = 3.71QRR329 pKa = 11.84NQSEE333 pKa = 4.18IQVASTVTT341 pKa = 3.13
Molecular weight: 37.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
649 |
308 |
341 |
324.5 |
36.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.553 ± 0.525 | 1.387 ± 0.374 |
5.547 ± 1.064 | 3.236 ± 0.873 |
4.777 ± 0.928 | 5.855 ± 0.44 |
3.544 ± 0.451 | 4.931 ± 0.609 |
5.085 ± 1.009 | 6.163 ± 0.437 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.849 ± 0.367 | 5.239 ± 0.895 |
4.931 ± 0.826 | 4.468 ± 0.268 |
5.547 ± 0.198 | 6.78 ± 1.706 |
7.088 ± 0.613 | 7.088 ± 1.262 |
2.619 ± 0.418 | 4.314 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
