
Hubei dimarhabdovirus virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
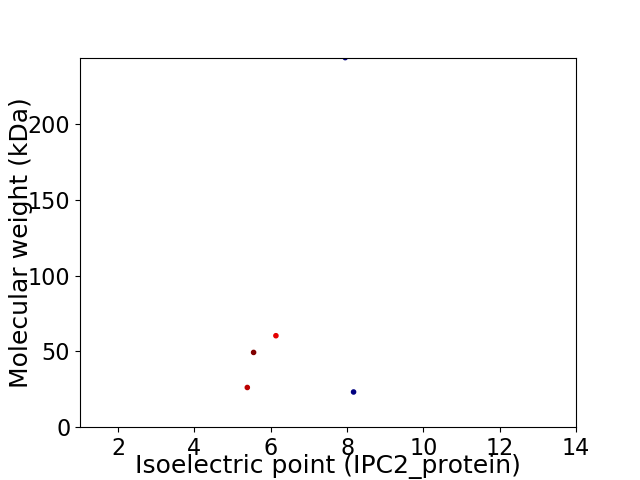
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMV4|A0A1L3KMV4_9VIRU Nucleocapsid protein OS=Hubei dimarhabdovirus virus 1 OX=1922866 PE=4 SV=1
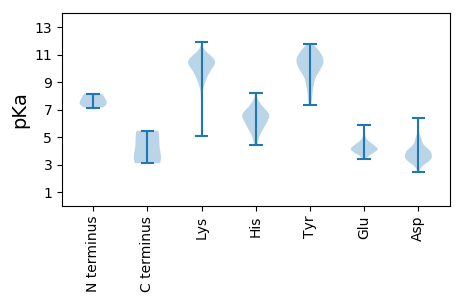
MM1 pKa = 7.9SFEE4 pKa = 4.41TEE6 pKa = 4.3SAHH9 pKa = 7.15HH10 pKa = 6.57VDD12 pKa = 3.97LAEE15 pKa = 4.38SLVKK19 pKa = 9.98TLTSMQKK26 pKa = 9.55NQTLSKK32 pKa = 10.86LPVFDD37 pKa = 5.82DD38 pKa = 3.86SSDD41 pKa = 3.57MTDD44 pKa = 3.42EE45 pKa = 4.4DD46 pKa = 4.86HH47 pKa = 7.77SNSDD51 pKa = 3.61PDD53 pKa = 3.88VVGAQYY59 pKa = 11.06LGDD62 pKa = 3.83PAEE65 pKa = 4.89LFGSLPPAVARR76 pKa = 11.84ATPQSLDD83 pKa = 3.47LSFLSLSDD91 pKa = 3.37AKK93 pKa = 10.4RR94 pKa = 11.84VQDD97 pKa = 5.25LICSLDD103 pKa = 3.28TSRR106 pKa = 11.84PIDD109 pKa = 4.16EE110 pKa = 4.97IEE112 pKa = 4.13FVLPSAPRR120 pKa = 11.84KK121 pKa = 9.56PSTPKK126 pKa = 9.83PSAPVVTEE134 pKa = 3.48QRR136 pKa = 11.84QPPKK140 pKa = 10.61ASSTSKK146 pKa = 9.44TRR148 pKa = 11.84MVKK151 pKa = 10.64LGILLEE157 pKa = 4.42KK158 pKa = 10.36GVEE161 pKa = 4.09VRR163 pKa = 11.84SRR165 pKa = 11.84YY166 pKa = 8.74PDD168 pKa = 3.44KK169 pKa = 11.15PPFKK173 pKa = 10.12IYY175 pKa = 10.55NGRR178 pKa = 11.84NGFSKK183 pKa = 10.35EE184 pKa = 3.69QLAARR189 pKa = 11.84YY190 pKa = 7.58QNRR193 pKa = 11.84EE194 pKa = 4.08LPEE197 pKa = 3.73TLYY200 pKa = 11.22DD201 pKa = 3.61VVKK204 pKa = 10.83EE205 pKa = 4.11IFTEE209 pKa = 4.22AKK211 pKa = 9.13NWGVFINRR219 pKa = 11.84YY220 pKa = 8.17HH221 pKa = 7.28AALLKK226 pKa = 10.59INLDD230 pKa = 3.67QYY232 pKa = 11.22SCEE235 pKa = 3.99
MM1 pKa = 7.9SFEE4 pKa = 4.41TEE6 pKa = 4.3SAHH9 pKa = 7.15HH10 pKa = 6.57VDD12 pKa = 3.97LAEE15 pKa = 4.38SLVKK19 pKa = 9.98TLTSMQKK26 pKa = 9.55NQTLSKK32 pKa = 10.86LPVFDD37 pKa = 5.82DD38 pKa = 3.86SSDD41 pKa = 3.57MTDD44 pKa = 3.42EE45 pKa = 4.4DD46 pKa = 4.86HH47 pKa = 7.77SNSDD51 pKa = 3.61PDD53 pKa = 3.88VVGAQYY59 pKa = 11.06LGDD62 pKa = 3.83PAEE65 pKa = 4.89LFGSLPPAVARR76 pKa = 11.84ATPQSLDD83 pKa = 3.47LSFLSLSDD91 pKa = 3.37AKK93 pKa = 10.4RR94 pKa = 11.84VQDD97 pKa = 5.25LICSLDD103 pKa = 3.28TSRR106 pKa = 11.84PIDD109 pKa = 4.16EE110 pKa = 4.97IEE112 pKa = 4.13FVLPSAPRR120 pKa = 11.84KK121 pKa = 9.56PSTPKK126 pKa = 9.83PSAPVVTEE134 pKa = 3.48QRR136 pKa = 11.84QPPKK140 pKa = 10.61ASSTSKK146 pKa = 9.44TRR148 pKa = 11.84MVKK151 pKa = 10.64LGILLEE157 pKa = 4.42KK158 pKa = 10.36GVEE161 pKa = 4.09VRR163 pKa = 11.84SRR165 pKa = 11.84YY166 pKa = 8.74PDD168 pKa = 3.44KK169 pKa = 11.15PPFKK173 pKa = 10.12IYY175 pKa = 10.55NGRR178 pKa = 11.84NGFSKK183 pKa = 10.35EE184 pKa = 3.69QLAARR189 pKa = 11.84YY190 pKa = 7.58QNRR193 pKa = 11.84EE194 pKa = 4.08LPEE197 pKa = 3.73TLYY200 pKa = 11.22DD201 pKa = 3.61VVKK204 pKa = 10.83EE205 pKa = 4.11IFTEE209 pKa = 4.22AKK211 pKa = 9.13NWGVFINRR219 pKa = 11.84YY220 pKa = 8.17HH221 pKa = 7.28AALLKK226 pKa = 10.59INLDD230 pKa = 3.67QYY232 pKa = 11.22SCEE235 pKa = 3.99
Molecular weight: 26.23 kDa
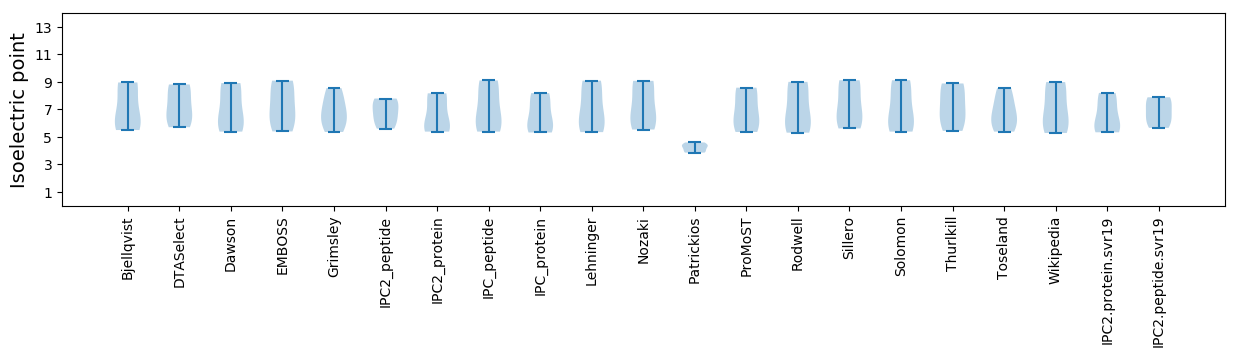
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMY8|A0A1L3KMY8_9VIRU GDP polyribonucleotidyltransferase OS=Hubei dimarhabdovirus virus 1 OX=1922866 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.13PITTALKK9 pKa = 10.2KK10 pKa = 10.32IRR12 pKa = 11.84GSPGAIRR19 pKa = 11.84SRR21 pKa = 11.84AQTTLPSLPPKK32 pKa = 10.37DD33 pKa = 3.98LRR35 pKa = 11.84CSKK38 pKa = 9.9WSIVAEE44 pKa = 4.16MKK46 pKa = 9.67LTLSQEE52 pKa = 4.01ALDD55 pKa = 3.73WEE57 pKa = 4.64IVQRR61 pKa = 11.84IAGVHH66 pKa = 5.5RR67 pKa = 11.84DD68 pKa = 3.67EE69 pKa = 4.63YY70 pKa = 10.71TGNPYY75 pKa = 9.79FRR77 pKa = 11.84PIIEE81 pKa = 4.04TFYY84 pKa = 10.54WLMLIHH90 pKa = 6.41TKK92 pKa = 10.4KK93 pKa = 10.94DD94 pKa = 3.1NTQITNSTTYY104 pKa = 10.71SSVFTEE110 pKa = 4.55LVSVHH115 pKa = 6.43HH116 pKa = 6.59NIPLVSNQDD125 pKa = 3.13FSYY128 pKa = 10.49SYY130 pKa = 9.12CTSGIHH136 pKa = 5.98RR137 pKa = 11.84HH138 pKa = 4.46TTFTIQFRR146 pKa = 11.84VKK148 pKa = 9.06FTPTVRR154 pKa = 11.84RR155 pKa = 11.84TEE157 pKa = 3.83DD158 pKa = 2.91LGFFLEE164 pKa = 4.6RR165 pKa = 11.84KK166 pKa = 8.32PFYY169 pKa = 10.16WSDD172 pKa = 3.77GPDD175 pKa = 3.36VIQVLKK181 pKa = 10.72MEE183 pKa = 4.75GLHH186 pKa = 6.87IALCGDD192 pKa = 4.22LLVIIDD198 pKa = 4.07PEE200 pKa = 4.32KK201 pKa = 10.9NN202 pKa = 3.1
MM1 pKa = 7.51KK2 pKa = 10.13PITTALKK9 pKa = 10.2KK10 pKa = 10.32IRR12 pKa = 11.84GSPGAIRR19 pKa = 11.84SRR21 pKa = 11.84AQTTLPSLPPKK32 pKa = 10.37DD33 pKa = 3.98LRR35 pKa = 11.84CSKK38 pKa = 9.9WSIVAEE44 pKa = 4.16MKK46 pKa = 9.67LTLSQEE52 pKa = 4.01ALDD55 pKa = 3.73WEE57 pKa = 4.64IVQRR61 pKa = 11.84IAGVHH66 pKa = 5.5RR67 pKa = 11.84DD68 pKa = 3.67EE69 pKa = 4.63YY70 pKa = 10.71TGNPYY75 pKa = 9.79FRR77 pKa = 11.84PIIEE81 pKa = 4.04TFYY84 pKa = 10.54WLMLIHH90 pKa = 6.41TKK92 pKa = 10.4KK93 pKa = 10.94DD94 pKa = 3.1NTQITNSTTYY104 pKa = 10.71SSVFTEE110 pKa = 4.55LVSVHH115 pKa = 6.43HH116 pKa = 6.59NIPLVSNQDD125 pKa = 3.13FSYY128 pKa = 10.49SYY130 pKa = 9.12CTSGIHH136 pKa = 5.98RR137 pKa = 11.84HH138 pKa = 4.46TTFTIQFRR146 pKa = 11.84VKK148 pKa = 9.06FTPTVRR154 pKa = 11.84RR155 pKa = 11.84TEE157 pKa = 3.83DD158 pKa = 2.91LGFFLEE164 pKa = 4.6RR165 pKa = 11.84KK166 pKa = 8.32PFYY169 pKa = 10.16WSDD172 pKa = 3.77GPDD175 pKa = 3.36VIQVLKK181 pKa = 10.72MEE183 pKa = 4.75GLHH186 pKa = 6.87IALCGDD192 pKa = 4.22LLVIIDD198 pKa = 4.07PEE200 pKa = 4.32KK201 pKa = 10.9NN202 pKa = 3.1
Molecular weight: 23.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3543 |
202 |
2135 |
708.6 |
80.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.403 ± 0.508 | 1.665 ± 0.16 |
5.588 ± 0.404 | 5.25 ± 0.175 |
4.601 ± 0.154 | 5.532 ± 0.55 |
2.653 ± 0.157 | 7.141 ± 0.645 |
6.125 ± 0.126 | 10.443 ± 0.5 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.258 | 5.25 ± 0.396 |
4.968 ± 0.696 | 3.189 ± 0.176 |
5.109 ± 0.472 | 7.988 ± 0.71 |
7.141 ± 0.474 | 5.306 ± 0.409 |
1.524 ± 0.143 | 3.98 ± 0.137 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
