
Ruminococcus sp. CAG:624
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; environmental samples
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
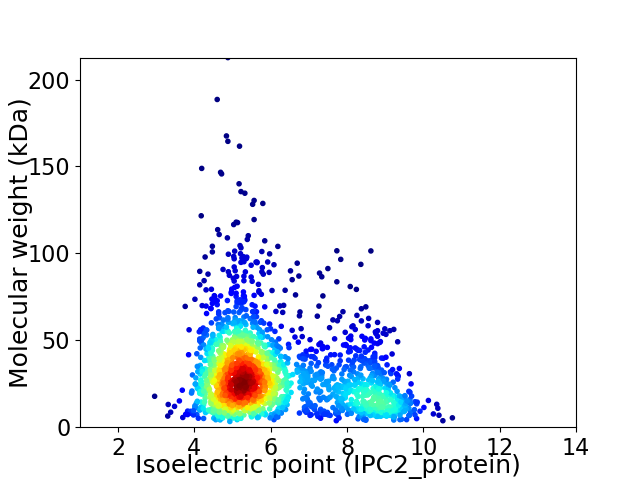
Virtual 2D-PAGE plot for 1963 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7MSH4|R7MSH4_9FIRM Malonyl CoA-acyl carrier protein transacylase OS=Ruminococcus sp. CAG:624 OX=1262965 GN=BN739_01946 PE=3 SV=1
MM1 pKa = 7.26NKK3 pKa = 9.73SRR5 pKa = 11.84ILTIILTALICVSAVSCNKK24 pKa = 9.97NSNKK28 pKa = 9.98SSEE31 pKa = 4.09NSSNEE36 pKa = 3.29ASQFATEE43 pKa = 4.21TTEE46 pKa = 3.98EE47 pKa = 4.26SSEE50 pKa = 4.06QTTEE54 pKa = 3.89EE55 pKa = 3.88QTDD58 pKa = 3.99TEE60 pKa = 4.38EE61 pKa = 4.35STDD64 pKa = 3.79SKK66 pKa = 11.54NEE68 pKa = 4.04SPDD71 pKa = 3.35NPFGKK76 pKa = 8.35TADD79 pKa = 3.37INDD82 pKa = 4.25YY83 pKa = 7.4FTPDD87 pKa = 3.37APEE90 pKa = 4.07PALWKK95 pKa = 9.61VTDD98 pKa = 4.37DD99 pKa = 3.92DD100 pKa = 4.68SGNVMYY106 pKa = 10.6MMGTIHH112 pKa = 6.32VASDD116 pKa = 3.35NSFSLPDD123 pKa = 4.09YY124 pKa = 11.42IMDD127 pKa = 4.36AYY129 pKa = 10.51EE130 pKa = 4.15NADD133 pKa = 3.51GVAVEE138 pKa = 3.92YY139 pKa = 10.56DD140 pKa = 3.38INKK143 pKa = 9.79LSSSLTSLALYY154 pKa = 10.51QNALLYY160 pKa = 10.67KK161 pKa = 10.62DD162 pKa = 3.23GTTVKK167 pKa = 10.53DD168 pKa = 3.59HH169 pKa = 6.96LSSEE173 pKa = 4.58TYY175 pKa = 10.15EE176 pKa = 3.95KK177 pKa = 10.93AKK179 pKa = 10.97NYY181 pKa = 10.04FDD183 pKa = 5.29DD184 pKa = 4.49LGVYY188 pKa = 10.02NEE190 pKa = 4.39QLDD193 pKa = 4.15KK194 pKa = 10.9YY195 pKa = 7.35TTGYY199 pKa = 9.7WIQQLNMIMFQRR211 pKa = 11.84MEE213 pKa = 4.24NLNLTGIDD221 pKa = 3.52SYY223 pKa = 11.62FIGLANKK230 pKa = 9.78DD231 pKa = 3.33GKK233 pKa = 10.32EE234 pKa = 3.97VVSIEE239 pKa = 4.33EE240 pKa = 3.96ISAQTDD246 pKa = 3.28ALNAYY251 pKa = 10.34SDD253 pKa = 4.43DD254 pKa = 3.76LADD257 pKa = 3.84YY258 pKa = 10.14VISDD262 pKa = 3.64MVDD265 pKa = 3.26NIDD268 pKa = 4.18EE269 pKa = 4.42IEE271 pKa = 4.66EE272 pKa = 4.25YY273 pKa = 8.36TQSVADD279 pKa = 5.22MYY281 pKa = 11.49NCWAKK286 pKa = 11.18GDD288 pKa = 3.67VDD290 pKa = 4.13KK291 pKa = 11.37LQEE294 pKa = 3.83IDD296 pKa = 3.47ADD298 pKa = 4.05EE299 pKa = 4.36MGEE302 pKa = 4.1LPKK305 pKa = 10.56EE306 pKa = 4.17LEE308 pKa = 4.22DD309 pKa = 6.49DD310 pKa = 3.86YY311 pKa = 11.94VQYY314 pKa = 11.44EE315 pKa = 4.19NIILHH320 pKa = 6.0NRR322 pKa = 11.84NQVMAQRR329 pKa = 11.84ASEE332 pKa = 4.39FIEE335 pKa = 4.61NNDD338 pKa = 3.26NLFYY342 pKa = 10.57MVGSLHH348 pKa = 6.41FAGDD352 pKa = 3.43QGVDD356 pKa = 3.9DD357 pKa = 5.1LLEE360 pKa = 4.81DD361 pKa = 3.01MGYY364 pKa = 7.96TVEE367 pKa = 4.69KK368 pKa = 10.58LHH370 pKa = 7.04
MM1 pKa = 7.26NKK3 pKa = 9.73SRR5 pKa = 11.84ILTIILTALICVSAVSCNKK24 pKa = 9.97NSNKK28 pKa = 9.98SSEE31 pKa = 4.09NSSNEE36 pKa = 3.29ASQFATEE43 pKa = 4.21TTEE46 pKa = 3.98EE47 pKa = 4.26SSEE50 pKa = 4.06QTTEE54 pKa = 3.89EE55 pKa = 3.88QTDD58 pKa = 3.99TEE60 pKa = 4.38EE61 pKa = 4.35STDD64 pKa = 3.79SKK66 pKa = 11.54NEE68 pKa = 4.04SPDD71 pKa = 3.35NPFGKK76 pKa = 8.35TADD79 pKa = 3.37INDD82 pKa = 4.25YY83 pKa = 7.4FTPDD87 pKa = 3.37APEE90 pKa = 4.07PALWKK95 pKa = 9.61VTDD98 pKa = 4.37DD99 pKa = 3.92DD100 pKa = 4.68SGNVMYY106 pKa = 10.6MMGTIHH112 pKa = 6.32VASDD116 pKa = 3.35NSFSLPDD123 pKa = 4.09YY124 pKa = 11.42IMDD127 pKa = 4.36AYY129 pKa = 10.51EE130 pKa = 4.15NADD133 pKa = 3.51GVAVEE138 pKa = 3.92YY139 pKa = 10.56DD140 pKa = 3.38INKK143 pKa = 9.79LSSSLTSLALYY154 pKa = 10.51QNALLYY160 pKa = 10.67KK161 pKa = 10.62DD162 pKa = 3.23GTTVKK167 pKa = 10.53DD168 pKa = 3.59HH169 pKa = 6.96LSSEE173 pKa = 4.58TYY175 pKa = 10.15EE176 pKa = 3.95KK177 pKa = 10.93AKK179 pKa = 10.97NYY181 pKa = 10.04FDD183 pKa = 5.29DD184 pKa = 4.49LGVYY188 pKa = 10.02NEE190 pKa = 4.39QLDD193 pKa = 4.15KK194 pKa = 10.9YY195 pKa = 7.35TTGYY199 pKa = 9.7WIQQLNMIMFQRR211 pKa = 11.84MEE213 pKa = 4.24NLNLTGIDD221 pKa = 3.52SYY223 pKa = 11.62FIGLANKK230 pKa = 9.78DD231 pKa = 3.33GKK233 pKa = 10.32EE234 pKa = 3.97VVSIEE239 pKa = 4.33EE240 pKa = 3.96ISAQTDD246 pKa = 3.28ALNAYY251 pKa = 10.34SDD253 pKa = 4.43DD254 pKa = 3.76LADD257 pKa = 3.84YY258 pKa = 10.14VISDD262 pKa = 3.64MVDD265 pKa = 3.26NIDD268 pKa = 4.18EE269 pKa = 4.42IEE271 pKa = 4.66EE272 pKa = 4.25YY273 pKa = 8.36TQSVADD279 pKa = 5.22MYY281 pKa = 11.49NCWAKK286 pKa = 11.18GDD288 pKa = 3.67VDD290 pKa = 4.13KK291 pKa = 11.37LQEE294 pKa = 3.83IDD296 pKa = 3.47ADD298 pKa = 4.05EE299 pKa = 4.36MGEE302 pKa = 4.1LPKK305 pKa = 10.56EE306 pKa = 4.17LEE308 pKa = 4.22DD309 pKa = 6.49DD310 pKa = 3.86YY311 pKa = 11.94VQYY314 pKa = 11.44EE315 pKa = 4.19NIILHH320 pKa = 6.0NRR322 pKa = 11.84NQVMAQRR329 pKa = 11.84ASEE332 pKa = 4.39FIEE335 pKa = 4.61NNDD338 pKa = 3.26NLFYY342 pKa = 10.57MVGSLHH348 pKa = 6.41FAGDD352 pKa = 3.43QGVDD356 pKa = 3.9DD357 pKa = 5.1LLEE360 pKa = 4.81DD361 pKa = 3.01MGYY364 pKa = 7.96TVEE367 pKa = 4.69KK368 pKa = 10.58LHH370 pKa = 7.04
Molecular weight: 41.69 kDa
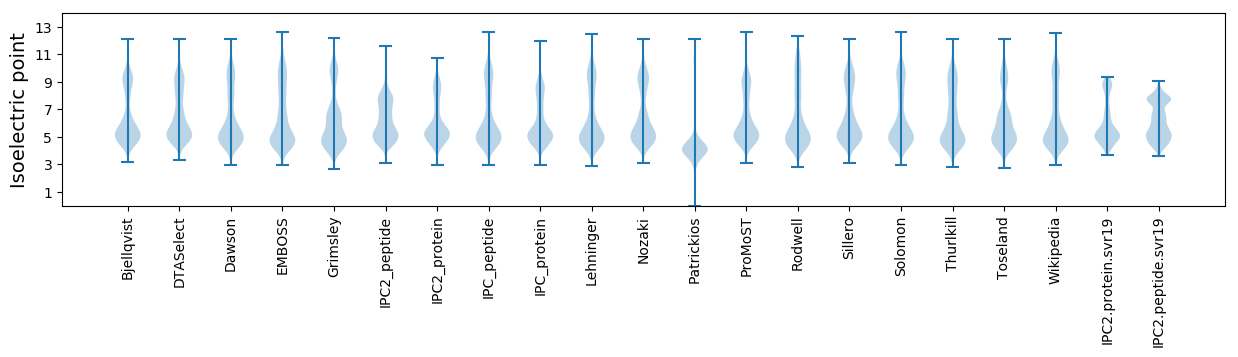
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7MQF4|R7MQF4_9FIRM 50S ribosomal protein L10 OS=Ruminococcus sp. CAG:624 OX=1262965 GN=rplJ PE=3 SV=1
MM1 pKa = 7.64KK2 pKa = 10.14YY3 pKa = 11.04VLIALIKK10 pKa = 10.17LYY12 pKa = 10.62RR13 pKa = 11.84KK14 pKa = 10.0LISPLFPPRR23 pKa = 11.84CKK25 pKa = 10.55YY26 pKa = 10.9YY27 pKa = 8.29PTCSSYY33 pKa = 11.48ALKK36 pKa = 10.55AVEE39 pKa = 3.69RR40 pKa = 11.84FGFFRR45 pKa = 11.84GTALAVWRR53 pKa = 11.84ILRR56 pKa = 11.84CNPWSLGGIDD66 pKa = 3.94PVPEE70 pKa = 3.69RR71 pKa = 11.84FSFKK75 pKa = 10.48RR76 pKa = 11.84NRR78 pKa = 3.34
MM1 pKa = 7.64KK2 pKa = 10.14YY3 pKa = 11.04VLIALIKK10 pKa = 10.17LYY12 pKa = 10.62RR13 pKa = 11.84KK14 pKa = 10.0LISPLFPPRR23 pKa = 11.84CKK25 pKa = 10.55YY26 pKa = 10.9YY27 pKa = 8.29PTCSSYY33 pKa = 11.48ALKK36 pKa = 10.55AVEE39 pKa = 3.69RR40 pKa = 11.84FGFFRR45 pKa = 11.84GTALAVWRR53 pKa = 11.84ILRR56 pKa = 11.84CNPWSLGGIDD66 pKa = 3.94PVPEE70 pKa = 3.69RR71 pKa = 11.84FSFKK75 pKa = 10.48RR76 pKa = 11.84NRR78 pKa = 3.34
Molecular weight: 9.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
571658 |
31 |
1910 |
291.2 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.989 ± 0.054 | 1.749 ± 0.023 |
5.979 ± 0.047 | 7.145 ± 0.059 |
4.371 ± 0.041 | 6.434 ± 0.05 |
1.515 ± 0.019 | 8.58 ± 0.048 |
7.599 ± 0.043 | 8.296 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.754 ± 0.028 | 5.546 ± 0.051 |
3.036 ± 0.03 | 2.557 ± 0.031 |
3.669 ± 0.042 | 7.096 ± 0.054 |
5.413 ± 0.046 | 6.39 ± 0.045 |
0.679 ± 0.016 | 4.182 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
