
Salinibacterium hongtaonis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Salinibacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
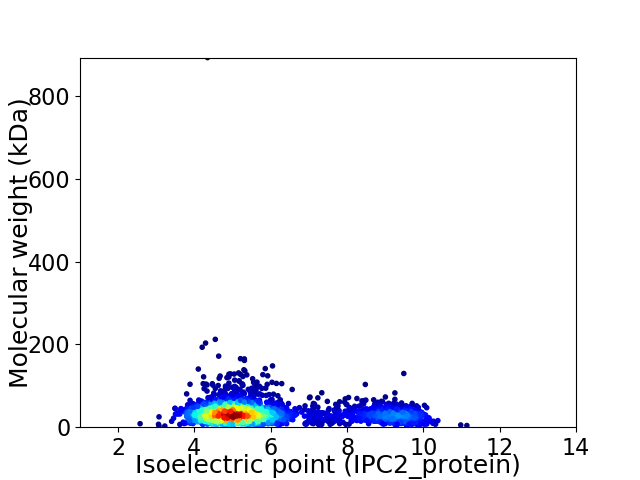
Virtual 2D-PAGE plot for 2605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U1T384|A0A2U1T384_9MICO MaoC family dehydratase OS=Salinibacterium hongtaonis OX=2079791 GN=DF220_11145 PE=3 SV=1
MM1 pKa = 7.39SRR3 pKa = 11.84LRR5 pKa = 11.84RR6 pKa = 11.84SAVALVAIIPLLLSGCFLIGGEE28 pKa = 4.35SRR30 pKa = 11.84TTSTPTGEE38 pKa = 4.66KK39 pKa = 10.13VDD41 pKa = 4.62GDD43 pKa = 3.92LQPFYY48 pKa = 10.87DD49 pKa = 4.98QVVEE53 pKa = 4.2WEE55 pKa = 4.23SCGDD59 pKa = 3.77DD60 pKa = 4.03FQCATAMAPLDD71 pKa = 3.41WDD73 pKa = 3.7DD74 pKa = 4.54PEE76 pKa = 5.85RR77 pKa = 11.84EE78 pKa = 4.32SIEE81 pKa = 3.93LALIRR86 pKa = 11.84LTTRR90 pKa = 11.84GTPMGSLLVNPGGPGGSGVDD110 pKa = 4.19FVRR113 pKa = 11.84EE114 pKa = 3.87SWDD117 pKa = 3.5YY118 pKa = 11.1ATTAQLRR125 pKa = 11.84DD126 pKa = 3.51NYY128 pKa = 10.86DD129 pKa = 3.12LVGFDD134 pKa = 3.8PRR136 pKa = 11.84GVGASSAVSCYY147 pKa = 10.5DD148 pKa = 4.78DD149 pKa = 4.91PDD151 pKa = 3.8TYY153 pKa = 11.37TDD155 pKa = 3.67YY156 pKa = 11.29LYY158 pKa = 10.38DD159 pKa = 3.75ITPGTPGSAEE169 pKa = 3.85WLDD172 pKa = 4.37AMDD175 pKa = 3.18ATMAEE180 pKa = 4.69FGADD184 pKa = 3.1CLEE187 pKa = 4.23FTGEE191 pKa = 4.01LLGFVDD197 pKa = 4.4TVSAARR203 pKa = 11.84DD204 pKa = 3.5LDD206 pKa = 3.97LLRR209 pKa = 11.84AILGDD214 pKa = 3.96DD215 pKa = 3.26KK216 pKa = 11.65LNYY219 pKa = 9.89LGSSYY224 pKa = 10.26GTFLGATYY232 pKa = 11.2ADD234 pKa = 5.52LYY236 pKa = 9.1PQNTGRR242 pKa = 11.84LVLDD246 pKa = 4.16GAIDD250 pKa = 3.72PASTDD255 pKa = 3.95FEE257 pKa = 4.62VTAMQAQGFEE267 pKa = 4.4SAMRR271 pKa = 11.84AYY273 pKa = 10.62LADD276 pKa = 4.14CLTGANCPFTGTVEE290 pKa = 4.4SGMKK294 pKa = 10.22EE295 pKa = 3.47MGAIFARR302 pKa = 11.84LEE304 pKa = 4.07ASPLRR309 pKa = 11.84AEE311 pKa = 4.91DD312 pKa = 3.61GRR314 pKa = 11.84LLGSQTMFTAVILPLYY330 pKa = 10.52SADD333 pKa = 2.59TWQYY337 pKa = 8.61LTSLITDD344 pKa = 3.92VKK346 pKa = 10.54RR347 pKa = 11.84GSAEE351 pKa = 3.8IAFSLADD358 pKa = 4.32LYY360 pKa = 11.34NGRR363 pKa = 11.84EE364 pKa = 4.04PDD366 pKa = 3.24GTFADD371 pKa = 3.87NSSEE375 pKa = 3.94AFTAINCLDD384 pKa = 3.85YY385 pKa = 10.83PSTSTRR391 pKa = 11.84EE392 pKa = 4.01TLPAEE397 pKa = 4.34AAEE400 pKa = 4.36LAALAPVFGPQMSWGGTSCDD420 pKa = 2.99AWPFEE425 pKa = 4.32ATRR428 pKa = 11.84VRR430 pKa = 11.84GPIAADD436 pKa = 4.19GSAPILVVGTTNDD449 pKa = 3.6PATPYY454 pKa = 10.0AWSVALADD462 pKa = 3.79QLQNGHH468 pKa = 6.31LVSYY472 pKa = 10.42DD473 pKa = 3.83GEE475 pKa = 4.3GHH477 pKa = 5.16TAYY480 pKa = 10.61NKK482 pKa = 10.43SNQCIDD488 pKa = 3.58DD489 pKa = 3.81TVDD492 pKa = 3.24AFFIDD497 pKa = 3.93GTVPATDD504 pKa = 4.05PLCC507 pKa = 4.35
MM1 pKa = 7.39SRR3 pKa = 11.84LRR5 pKa = 11.84RR6 pKa = 11.84SAVALVAIIPLLLSGCFLIGGEE28 pKa = 4.35SRR30 pKa = 11.84TTSTPTGEE38 pKa = 4.66KK39 pKa = 10.13VDD41 pKa = 4.62GDD43 pKa = 3.92LQPFYY48 pKa = 10.87DD49 pKa = 4.98QVVEE53 pKa = 4.2WEE55 pKa = 4.23SCGDD59 pKa = 3.77DD60 pKa = 4.03FQCATAMAPLDD71 pKa = 3.41WDD73 pKa = 3.7DD74 pKa = 4.54PEE76 pKa = 5.85RR77 pKa = 11.84EE78 pKa = 4.32SIEE81 pKa = 3.93LALIRR86 pKa = 11.84LTTRR90 pKa = 11.84GTPMGSLLVNPGGPGGSGVDD110 pKa = 4.19FVRR113 pKa = 11.84EE114 pKa = 3.87SWDD117 pKa = 3.5YY118 pKa = 11.1ATTAQLRR125 pKa = 11.84DD126 pKa = 3.51NYY128 pKa = 10.86DD129 pKa = 3.12LVGFDD134 pKa = 3.8PRR136 pKa = 11.84GVGASSAVSCYY147 pKa = 10.5DD148 pKa = 4.78DD149 pKa = 4.91PDD151 pKa = 3.8TYY153 pKa = 11.37TDD155 pKa = 3.67YY156 pKa = 11.29LYY158 pKa = 10.38DD159 pKa = 3.75ITPGTPGSAEE169 pKa = 3.85WLDD172 pKa = 4.37AMDD175 pKa = 3.18ATMAEE180 pKa = 4.69FGADD184 pKa = 3.1CLEE187 pKa = 4.23FTGEE191 pKa = 4.01LLGFVDD197 pKa = 4.4TVSAARR203 pKa = 11.84DD204 pKa = 3.5LDD206 pKa = 3.97LLRR209 pKa = 11.84AILGDD214 pKa = 3.96DD215 pKa = 3.26KK216 pKa = 11.65LNYY219 pKa = 9.89LGSSYY224 pKa = 10.26GTFLGATYY232 pKa = 11.2ADD234 pKa = 5.52LYY236 pKa = 9.1PQNTGRR242 pKa = 11.84LVLDD246 pKa = 4.16GAIDD250 pKa = 3.72PASTDD255 pKa = 3.95FEE257 pKa = 4.62VTAMQAQGFEE267 pKa = 4.4SAMRR271 pKa = 11.84AYY273 pKa = 10.62LADD276 pKa = 4.14CLTGANCPFTGTVEE290 pKa = 4.4SGMKK294 pKa = 10.22EE295 pKa = 3.47MGAIFARR302 pKa = 11.84LEE304 pKa = 4.07ASPLRR309 pKa = 11.84AEE311 pKa = 4.91DD312 pKa = 3.61GRR314 pKa = 11.84LLGSQTMFTAVILPLYY330 pKa = 10.52SADD333 pKa = 2.59TWQYY337 pKa = 8.61LTSLITDD344 pKa = 3.92VKK346 pKa = 10.54RR347 pKa = 11.84GSAEE351 pKa = 3.8IAFSLADD358 pKa = 4.32LYY360 pKa = 11.34NGRR363 pKa = 11.84EE364 pKa = 4.04PDD366 pKa = 3.24GTFADD371 pKa = 3.87NSSEE375 pKa = 3.94AFTAINCLDD384 pKa = 3.85YY385 pKa = 10.83PSTSTRR391 pKa = 11.84EE392 pKa = 4.01TLPAEE397 pKa = 4.34AAEE400 pKa = 4.36LAALAPVFGPQMSWGGTSCDD420 pKa = 2.99AWPFEE425 pKa = 4.32ATRR428 pKa = 11.84VRR430 pKa = 11.84GPIAADD436 pKa = 4.19GSAPILVVGTTNDD449 pKa = 3.6PATPYY454 pKa = 10.0AWSVALADD462 pKa = 3.79QLQNGHH468 pKa = 6.31LVSYY472 pKa = 10.42DD473 pKa = 3.83GEE475 pKa = 4.3GHH477 pKa = 5.16TAYY480 pKa = 10.61NKK482 pKa = 10.43SNQCIDD488 pKa = 3.58DD489 pKa = 3.81TVDD492 pKa = 3.24AFFIDD497 pKa = 3.93GTVPATDD504 pKa = 4.05PLCC507 pKa = 4.35
Molecular weight: 54.09 kDa
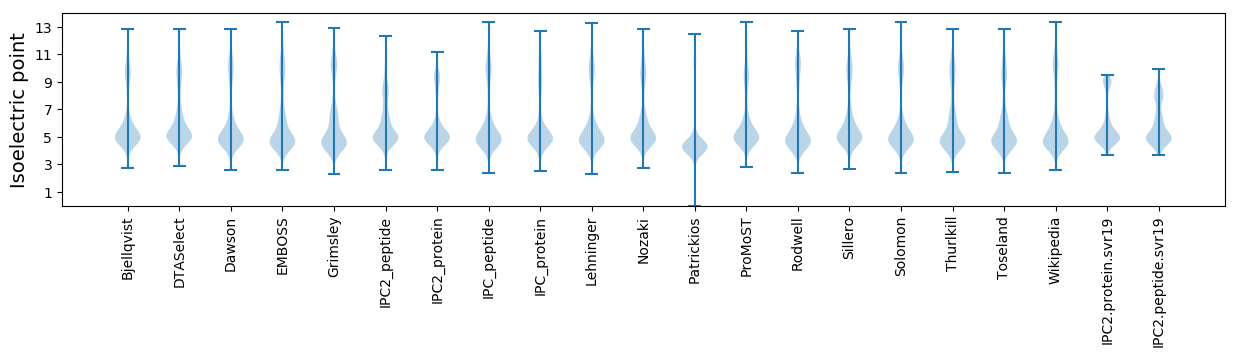
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U1T2F2|A0A2U1T2F2_9MICO Maltose ABC transporter permease OS=Salinibacterium hongtaonis OX=2079791 GN=DF220_09710 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
856357 |
24 |
9317 |
328.7 |
35.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.003 ± 0.083 | 0.483 ± 0.01 |
5.855 ± 0.041 | 5.723 ± 0.049 |
3.241 ± 0.037 | 8.752 ± 0.059 |
1.907 ± 0.022 | 5.145 ± 0.033 |
2.257 ± 0.036 | 9.997 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.927 ± 0.025 | 2.277 ± 0.029 |
5.104 ± 0.041 | 2.842 ± 0.026 |
6.619 ± 0.062 | 6.453 ± 0.045 |
6.142 ± 0.079 | 8.923 ± 0.042 |
1.369 ± 0.021 | 1.981 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
