
Bacteroidales bacterium 6E
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; unclassified Bacteroidales
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
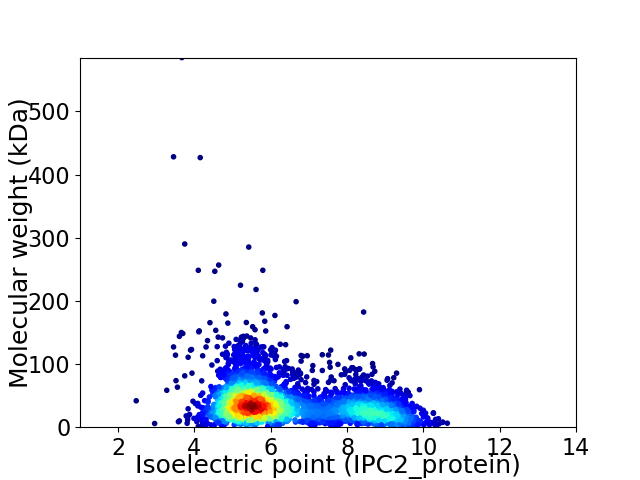
Virtual 2D-PAGE plot for 3618 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K8R1Y1|A0A0K8R1Y1_9BACT Uncharacterized protein OS=Bacteroidales bacterium 6E OX=1688776 GN=BA6E_12540 PE=4 SV=1
MM1 pKa = 7.28EE2 pKa = 5.24ASSRR6 pKa = 11.84NWAPASSYY14 pKa = 11.47SEE16 pKa = 4.13IIKK19 pKa = 10.43QNTMNKK25 pKa = 9.3FLLFVAMILSFGHH38 pKa = 6.5TMAQDD43 pKa = 3.65HH44 pKa = 6.58FKK46 pKa = 10.83VAYY49 pKa = 8.61TEE51 pKa = 4.14QPQTYY56 pKa = 8.16MNIYY60 pKa = 9.33VFSASVGGNNLGAGDD75 pKa = 4.85EE76 pKa = 4.17IAVFDD81 pKa = 3.87GTICCGVYY89 pKa = 10.07KK90 pKa = 10.8VSGAFAGPVNIPAAKK105 pKa = 10.29AEE107 pKa = 4.11AAGTGYY113 pKa = 8.19TAGNPIVVKK122 pKa = 10.45IWDD125 pKa = 3.46ASAGIEE131 pKa = 4.01YY132 pKa = 10.6SADD135 pKa = 2.66ISFRR139 pKa = 11.84ANSPNTVFTNNEE151 pKa = 3.85SAYY154 pKa = 9.25MDD156 pKa = 3.59ISAKK160 pKa = 9.81VAATIQLTANNKK172 pKa = 9.08TYY174 pKa = 10.99DD175 pKa = 3.45GTTNADD181 pKa = 2.7VGYY184 pKa = 10.09SVVDD188 pKa = 4.1GSLAGDD194 pKa = 3.23ITVNVANGLFADD206 pKa = 4.1KK207 pKa = 10.53HH208 pKa = 6.22AGSGKK213 pKa = 9.61PVTADD218 pKa = 3.0ITVTGADD225 pKa = 3.65IEE227 pKa = 4.78NFDD230 pKa = 3.82LTIIEE235 pKa = 4.56TTTATISKK243 pKa = 8.55KK244 pKa = 7.49TVEE247 pKa = 4.67VINAVAQDD255 pKa = 3.37KK256 pKa = 10.66VYY258 pKa = 11.05NGNAVAVVSGASLAGVITPDD278 pKa = 3.15EE279 pKa = 4.5VIISGGTSGTFAQSNVGNAISVTTAMALTGSGASNYY315 pKa = 7.55EE316 pKa = 4.39LKK318 pKa = 10.73SQPALSASITKK329 pKa = 10.56APLTVTAEE337 pKa = 4.45NKK339 pKa = 10.24SKK341 pKa = 9.65TYY343 pKa = 10.51DD344 pKa = 3.42GQVYY348 pKa = 10.65SGFTSKK354 pKa = 8.83ITGFVGGEE362 pKa = 4.13TQSVISGSVTFTGDD376 pKa = 3.33AISAINAGSYY386 pKa = 8.31TVSPVVAGLSATNYY400 pKa = 10.35SFSTANGTLTVSKK413 pKa = 11.05APLTVTANDD422 pKa = 3.23QTKK425 pKa = 10.27VYY427 pKa = 10.62GSANPALTIAYY438 pKa = 9.56SGFVNGEE445 pKa = 4.14TASVLATAPTASTQINATTNAGTYY469 pKa = 10.31ADD471 pKa = 5.58AITISGGVDD480 pKa = 3.03EE481 pKa = 5.75NYY483 pKa = 10.48SFSYY487 pKa = 10.21VAGDD491 pKa = 3.4FTVTKK496 pKa = 10.78APLTVTANDD505 pKa = 3.23QTKK508 pKa = 10.27VYY510 pKa = 10.62GSANPALTIAYY521 pKa = 9.9SGFEE525 pKa = 4.06NGEE528 pKa = 4.17TAQVLATAPTATTQISATTSAGTYY552 pKa = 8.61TDD554 pKa = 5.41AITVSGGVDD563 pKa = 3.22EE564 pKa = 5.56NYY566 pKa = 10.34SFSYY570 pKa = 10.17IAGDD574 pKa = 3.55FTVTKK579 pKa = 10.79APLTVVANDD588 pKa = 3.37QIKK591 pKa = 10.69VYY593 pKa = 10.75GSANPVLTFNYY604 pKa = 9.99SGFVNGEE611 pKa = 4.1TAPVLATAPSAATLINATTNAGTYY635 pKa = 9.8VDD637 pKa = 5.29AISVSGGVDD646 pKa = 3.13EE647 pKa = 5.63NYY649 pKa = 10.71SFTYY653 pKa = 9.86VAGDD657 pKa = 3.39FTVTKK662 pKa = 8.81ATPAIQVTGGSYY674 pKa = 10.92VFDD677 pKa = 3.85GTGKK681 pKa = 10.12SATAFAYY688 pKa = 10.07GVGGTSDD695 pKa = 3.69VLSPAVTITYY705 pKa = 8.76TGADD709 pKa = 3.49GTIYY713 pKa = 10.55GPSNIAPSASGSYY726 pKa = 7.17QVKK729 pKa = 10.61ADD731 pKa = 3.77FVGNTNYY738 pKa = 10.65LAISATASLIIANTVSTITVTGPNSYY764 pKa = 9.26TYY766 pKa = 10.53NGQAQGPAAAEE777 pKa = 4.3VTGSTGAVTYY787 pKa = 9.64IYY789 pKa = 10.37TGIAGTNYY797 pKa = 9.64GPSSTPPMNAGSYY810 pKa = 8.27QVVATVAADD819 pKa = 3.38ATYY822 pKa = 10.72QGASSAAFVFAIQKK836 pKa = 10.61ASLTITATSQGRR848 pKa = 11.84VYY850 pKa = 10.99GIDD853 pKa = 3.52NPQLTLSFEE862 pKa = 4.4GFVGGDD868 pKa = 3.63DD869 pKa = 3.31EE870 pKa = 4.79TVIDD874 pKa = 4.61EE875 pKa = 5.05LPLASTDD882 pKa = 3.69AVISSDD888 pKa = 2.8AGAYY892 pKa = 9.57IIMVSGGNDD901 pKa = 2.82NNYY904 pKa = 9.69QFIYY908 pKa = 10.51KK909 pKa = 10.04EE910 pKa = 4.16GVLTIMKK917 pKa = 9.81AQATIQLLNLEE928 pKa = 4.15QAYY931 pKa = 8.8TGKK934 pKa = 9.4PLEE937 pKa = 4.32VSYY940 pKa = 8.99QTTPAGLLVLFQYY953 pKa = 10.94DD954 pKa = 4.91GINILPSEE962 pKa = 4.23LGEE965 pKa = 4.21YY966 pKa = 10.32VVTAVVSDD974 pKa = 3.94KK975 pKa = 10.9NHH977 pKa = 6.37EE978 pKa = 4.22GSTTGIFRR986 pKa = 11.84ILEE989 pKa = 4.43DD990 pKa = 4.06EE991 pKa = 5.22DD992 pKa = 5.79GDD994 pKa = 4.69GIPDD998 pKa = 4.01IYY1000 pKa = 11.46DD1001 pKa = 3.8LDD1003 pKa = 4.08SDD1005 pKa = 4.03NDD1007 pKa = 4.04GVPDD1011 pKa = 3.87YY1012 pKa = 11.17LDD1014 pKa = 4.32KK1015 pKa = 11.24FPEE1018 pKa = 5.08DD1019 pKa = 3.68SNEE1022 pKa = 3.93WADD1025 pKa = 4.0SDD1027 pKa = 4.76GDD1029 pKa = 4.44GIGDD1033 pKa = 4.38NADD1036 pKa = 3.52TDD1038 pKa = 4.17DD1039 pKa = 5.88DD1040 pKa = 4.65NDD1042 pKa = 4.17GVDD1045 pKa = 4.48DD1046 pKa = 5.02VDD1048 pKa = 5.1DD1049 pKa = 5.24AFPNDD1054 pKa = 3.59PDD1056 pKa = 3.27EE1057 pKa = 4.83WEE1059 pKa = 5.11DD1060 pKa = 4.01SDD1062 pKa = 5.78GDD1064 pKa = 4.46GIGNNADD1071 pKa = 3.71SDD1073 pKa = 4.19DD1074 pKa = 5.15DD1075 pKa = 4.94GDD1077 pKa = 4.27GVADD1081 pKa = 4.34EE1082 pKa = 5.82DD1083 pKa = 5.45DD1084 pKa = 4.81AFPLNPDD1091 pKa = 2.87EE1092 pKa = 4.8WLDD1095 pKa = 3.79SDD1097 pKa = 5.0GDD1099 pKa = 4.46GIGDD1103 pKa = 4.38NADD1106 pKa = 3.52TDD1108 pKa = 4.29DD1109 pKa = 5.89DD1110 pKa = 4.67NDD1112 pKa = 4.07GTPDD1116 pKa = 3.51EE1117 pKa = 4.33QDD1119 pKa = 3.69AFPFDD1124 pKa = 4.55PNEE1127 pKa = 4.47DD1128 pKa = 3.65TDD1130 pKa = 4.15SDD1132 pKa = 4.2GDD1134 pKa = 4.29GVGDD1138 pKa = 4.15NSDD1141 pKa = 4.89LFPLDD1146 pKa = 4.36PEE1148 pKa = 4.25QSDD1151 pKa = 3.84DD1152 pKa = 3.81QVAPEE1157 pKa = 4.61LDD1159 pKa = 3.81VTQLTPLTLVCGIDD1173 pKa = 4.2FEE1175 pKa = 4.61EE1176 pKa = 5.57QLIAWLDD1183 pKa = 3.64SQAGVVATDD1192 pKa = 2.99NHH1194 pKa = 6.34QLDD1197 pKa = 4.27AWSHH1201 pKa = 6.03DD1202 pKa = 3.64LDD1204 pKa = 3.94IEE1206 pKa = 4.66EE1207 pKa = 5.11ILSDD1211 pKa = 4.18LCNEE1215 pKa = 4.37SKK1217 pKa = 9.94TIPVEE1222 pKa = 4.23FTVTDD1227 pKa = 3.53YY1228 pKa = 11.58SGNSSSFTATLVVVVNAAPEE1248 pKa = 4.38VISPLEE1254 pKa = 4.08DD1255 pKa = 2.74VVMAVNATHH1264 pKa = 7.62RR1265 pKa = 11.84IFASPLPGEE1274 pKa = 4.5MFGDD1278 pKa = 4.19DD1279 pKa = 3.22QDD1281 pKa = 3.49QALEE1285 pKa = 3.97VRR1287 pKa = 11.84YY1288 pKa = 8.63FALGSDD1294 pKa = 4.39TLPSWAKK1301 pKa = 10.07VVNDD1305 pKa = 3.77TLVFSPAIADD1315 pKa = 3.61TGCYY1319 pKa = 9.25QLVVEE1324 pKa = 6.1ARR1326 pKa = 11.84DD1327 pKa = 2.97IWQASVTDD1335 pKa = 3.93TFQVCIRR1342 pKa = 11.84ATLVNSDD1349 pKa = 4.06PLTMNGHH1356 pKa = 7.01GIIVYY1361 pKa = 7.95PNPASDD1367 pKa = 4.24FVRR1370 pKa = 11.84IKK1372 pKa = 10.34PGKK1375 pKa = 8.82QPEE1378 pKa = 4.48TQVEE1382 pKa = 4.51VTLMNLSGQAVIHH1395 pKa = 5.55RR1396 pKa = 11.84QFWSDD1401 pKa = 3.46EE1402 pKa = 3.91PLEE1405 pKa = 5.49IDD1407 pKa = 4.23LTGQKK1412 pKa = 9.31TGMYY1416 pKa = 9.59VIRR1419 pKa = 11.84ITFNNQTMYY1428 pKa = 10.75NKK1430 pKa = 10.55LMIEE1434 pKa = 4.11RR1435 pKa = 11.84RR1436 pKa = 3.69
MM1 pKa = 7.28EE2 pKa = 5.24ASSRR6 pKa = 11.84NWAPASSYY14 pKa = 11.47SEE16 pKa = 4.13IIKK19 pKa = 10.43QNTMNKK25 pKa = 9.3FLLFVAMILSFGHH38 pKa = 6.5TMAQDD43 pKa = 3.65HH44 pKa = 6.58FKK46 pKa = 10.83VAYY49 pKa = 8.61TEE51 pKa = 4.14QPQTYY56 pKa = 8.16MNIYY60 pKa = 9.33VFSASVGGNNLGAGDD75 pKa = 4.85EE76 pKa = 4.17IAVFDD81 pKa = 3.87GTICCGVYY89 pKa = 10.07KK90 pKa = 10.8VSGAFAGPVNIPAAKK105 pKa = 10.29AEE107 pKa = 4.11AAGTGYY113 pKa = 8.19TAGNPIVVKK122 pKa = 10.45IWDD125 pKa = 3.46ASAGIEE131 pKa = 4.01YY132 pKa = 10.6SADD135 pKa = 2.66ISFRR139 pKa = 11.84ANSPNTVFTNNEE151 pKa = 3.85SAYY154 pKa = 9.25MDD156 pKa = 3.59ISAKK160 pKa = 9.81VAATIQLTANNKK172 pKa = 9.08TYY174 pKa = 10.99DD175 pKa = 3.45GTTNADD181 pKa = 2.7VGYY184 pKa = 10.09SVVDD188 pKa = 4.1GSLAGDD194 pKa = 3.23ITVNVANGLFADD206 pKa = 4.1KK207 pKa = 10.53HH208 pKa = 6.22AGSGKK213 pKa = 9.61PVTADD218 pKa = 3.0ITVTGADD225 pKa = 3.65IEE227 pKa = 4.78NFDD230 pKa = 3.82LTIIEE235 pKa = 4.56TTTATISKK243 pKa = 8.55KK244 pKa = 7.49TVEE247 pKa = 4.67VINAVAQDD255 pKa = 3.37KK256 pKa = 10.66VYY258 pKa = 11.05NGNAVAVVSGASLAGVITPDD278 pKa = 3.15EE279 pKa = 4.5VIISGGTSGTFAQSNVGNAISVTTAMALTGSGASNYY315 pKa = 7.55EE316 pKa = 4.39LKK318 pKa = 10.73SQPALSASITKK329 pKa = 10.56APLTVTAEE337 pKa = 4.45NKK339 pKa = 10.24SKK341 pKa = 9.65TYY343 pKa = 10.51DD344 pKa = 3.42GQVYY348 pKa = 10.65SGFTSKK354 pKa = 8.83ITGFVGGEE362 pKa = 4.13TQSVISGSVTFTGDD376 pKa = 3.33AISAINAGSYY386 pKa = 8.31TVSPVVAGLSATNYY400 pKa = 10.35SFSTANGTLTVSKK413 pKa = 11.05APLTVTANDD422 pKa = 3.23QTKK425 pKa = 10.27VYY427 pKa = 10.62GSANPALTIAYY438 pKa = 9.56SGFVNGEE445 pKa = 4.14TASVLATAPTASTQINATTNAGTYY469 pKa = 10.31ADD471 pKa = 5.58AITISGGVDD480 pKa = 3.03EE481 pKa = 5.75NYY483 pKa = 10.48SFSYY487 pKa = 10.21VAGDD491 pKa = 3.4FTVTKK496 pKa = 10.78APLTVTANDD505 pKa = 3.23QTKK508 pKa = 10.27VYY510 pKa = 10.62GSANPALTIAYY521 pKa = 9.9SGFEE525 pKa = 4.06NGEE528 pKa = 4.17TAQVLATAPTATTQISATTSAGTYY552 pKa = 8.61TDD554 pKa = 5.41AITVSGGVDD563 pKa = 3.22EE564 pKa = 5.56NYY566 pKa = 10.34SFSYY570 pKa = 10.17IAGDD574 pKa = 3.55FTVTKK579 pKa = 10.79APLTVVANDD588 pKa = 3.37QIKK591 pKa = 10.69VYY593 pKa = 10.75GSANPVLTFNYY604 pKa = 9.99SGFVNGEE611 pKa = 4.1TAPVLATAPSAATLINATTNAGTYY635 pKa = 9.8VDD637 pKa = 5.29AISVSGGVDD646 pKa = 3.13EE647 pKa = 5.63NYY649 pKa = 10.71SFTYY653 pKa = 9.86VAGDD657 pKa = 3.39FTVTKK662 pKa = 8.81ATPAIQVTGGSYY674 pKa = 10.92VFDD677 pKa = 3.85GTGKK681 pKa = 10.12SATAFAYY688 pKa = 10.07GVGGTSDD695 pKa = 3.69VLSPAVTITYY705 pKa = 8.76TGADD709 pKa = 3.49GTIYY713 pKa = 10.55GPSNIAPSASGSYY726 pKa = 7.17QVKK729 pKa = 10.61ADD731 pKa = 3.77FVGNTNYY738 pKa = 10.65LAISATASLIIANTVSTITVTGPNSYY764 pKa = 9.26TYY766 pKa = 10.53NGQAQGPAAAEE777 pKa = 4.3VTGSTGAVTYY787 pKa = 9.64IYY789 pKa = 10.37TGIAGTNYY797 pKa = 9.64GPSSTPPMNAGSYY810 pKa = 8.27QVVATVAADD819 pKa = 3.38ATYY822 pKa = 10.72QGASSAAFVFAIQKK836 pKa = 10.61ASLTITATSQGRR848 pKa = 11.84VYY850 pKa = 10.99GIDD853 pKa = 3.52NPQLTLSFEE862 pKa = 4.4GFVGGDD868 pKa = 3.63DD869 pKa = 3.31EE870 pKa = 4.79TVIDD874 pKa = 4.61EE875 pKa = 5.05LPLASTDD882 pKa = 3.69AVISSDD888 pKa = 2.8AGAYY892 pKa = 9.57IIMVSGGNDD901 pKa = 2.82NNYY904 pKa = 9.69QFIYY908 pKa = 10.51KK909 pKa = 10.04EE910 pKa = 4.16GVLTIMKK917 pKa = 9.81AQATIQLLNLEE928 pKa = 4.15QAYY931 pKa = 8.8TGKK934 pKa = 9.4PLEE937 pKa = 4.32VSYY940 pKa = 8.99QTTPAGLLVLFQYY953 pKa = 10.94DD954 pKa = 4.91GINILPSEE962 pKa = 4.23LGEE965 pKa = 4.21YY966 pKa = 10.32VVTAVVSDD974 pKa = 3.94KK975 pKa = 10.9NHH977 pKa = 6.37EE978 pKa = 4.22GSTTGIFRR986 pKa = 11.84ILEE989 pKa = 4.43DD990 pKa = 4.06EE991 pKa = 5.22DD992 pKa = 5.79GDD994 pKa = 4.69GIPDD998 pKa = 4.01IYY1000 pKa = 11.46DD1001 pKa = 3.8LDD1003 pKa = 4.08SDD1005 pKa = 4.03NDD1007 pKa = 4.04GVPDD1011 pKa = 3.87YY1012 pKa = 11.17LDD1014 pKa = 4.32KK1015 pKa = 11.24FPEE1018 pKa = 5.08DD1019 pKa = 3.68SNEE1022 pKa = 3.93WADD1025 pKa = 4.0SDD1027 pKa = 4.76GDD1029 pKa = 4.44GIGDD1033 pKa = 4.38NADD1036 pKa = 3.52TDD1038 pKa = 4.17DD1039 pKa = 5.88DD1040 pKa = 4.65NDD1042 pKa = 4.17GVDD1045 pKa = 4.48DD1046 pKa = 5.02VDD1048 pKa = 5.1DD1049 pKa = 5.24AFPNDD1054 pKa = 3.59PDD1056 pKa = 3.27EE1057 pKa = 4.83WEE1059 pKa = 5.11DD1060 pKa = 4.01SDD1062 pKa = 5.78GDD1064 pKa = 4.46GIGNNADD1071 pKa = 3.71SDD1073 pKa = 4.19DD1074 pKa = 5.15DD1075 pKa = 4.94GDD1077 pKa = 4.27GVADD1081 pKa = 4.34EE1082 pKa = 5.82DD1083 pKa = 5.45DD1084 pKa = 4.81AFPLNPDD1091 pKa = 2.87EE1092 pKa = 4.8WLDD1095 pKa = 3.79SDD1097 pKa = 5.0GDD1099 pKa = 4.46GIGDD1103 pKa = 4.38NADD1106 pKa = 3.52TDD1108 pKa = 4.29DD1109 pKa = 5.89DD1110 pKa = 4.67NDD1112 pKa = 4.07GTPDD1116 pKa = 3.51EE1117 pKa = 4.33QDD1119 pKa = 3.69AFPFDD1124 pKa = 4.55PNEE1127 pKa = 4.47DD1128 pKa = 3.65TDD1130 pKa = 4.15SDD1132 pKa = 4.2GDD1134 pKa = 4.29GVGDD1138 pKa = 4.15NSDD1141 pKa = 4.89LFPLDD1146 pKa = 4.36PEE1148 pKa = 4.25QSDD1151 pKa = 3.84DD1152 pKa = 3.81QVAPEE1157 pKa = 4.61LDD1159 pKa = 3.81VTQLTPLTLVCGIDD1173 pKa = 4.2FEE1175 pKa = 4.61EE1176 pKa = 5.57QLIAWLDD1183 pKa = 3.64SQAGVVATDD1192 pKa = 2.99NHH1194 pKa = 6.34QLDD1197 pKa = 4.27AWSHH1201 pKa = 6.03DD1202 pKa = 3.64LDD1204 pKa = 3.94IEE1206 pKa = 4.66EE1207 pKa = 5.11ILSDD1211 pKa = 4.18LCNEE1215 pKa = 4.37SKK1217 pKa = 9.94TIPVEE1222 pKa = 4.23FTVTDD1227 pKa = 3.53YY1228 pKa = 11.58SGNSSSFTATLVVVVNAAPEE1248 pKa = 4.38VISPLEE1254 pKa = 4.08DD1255 pKa = 2.74VVMAVNATHH1264 pKa = 7.62RR1265 pKa = 11.84IFASPLPGEE1274 pKa = 4.5MFGDD1278 pKa = 4.19DD1279 pKa = 3.22QDD1281 pKa = 3.49QALEE1285 pKa = 3.97VRR1287 pKa = 11.84YY1288 pKa = 8.63FALGSDD1294 pKa = 4.39TLPSWAKK1301 pKa = 10.07VVNDD1305 pKa = 3.77TLVFSPAIADD1315 pKa = 3.61TGCYY1319 pKa = 9.25QLVVEE1324 pKa = 6.1ARR1326 pKa = 11.84DD1327 pKa = 2.97IWQASVTDD1335 pKa = 3.93TFQVCIRR1342 pKa = 11.84ATLVNSDD1349 pKa = 4.06PLTMNGHH1356 pKa = 7.01GIIVYY1361 pKa = 7.95PNPASDD1367 pKa = 4.24FVRR1370 pKa = 11.84IKK1372 pKa = 10.34PGKK1375 pKa = 8.82QPEE1378 pKa = 4.48TQVEE1382 pKa = 4.51VTLMNLSGQAVIHH1395 pKa = 5.55RR1396 pKa = 11.84QFWSDD1401 pKa = 3.46EE1402 pKa = 3.91PLEE1405 pKa = 5.49IDD1407 pKa = 4.23LTGQKK1412 pKa = 9.31TGMYY1416 pKa = 9.59VIRR1419 pKa = 11.84ITFNNQTMYY1428 pKa = 10.75NKK1430 pKa = 10.55LMIEE1434 pKa = 4.11RR1435 pKa = 11.84RR1436 pKa = 3.69
Molecular weight: 149.84 kDa
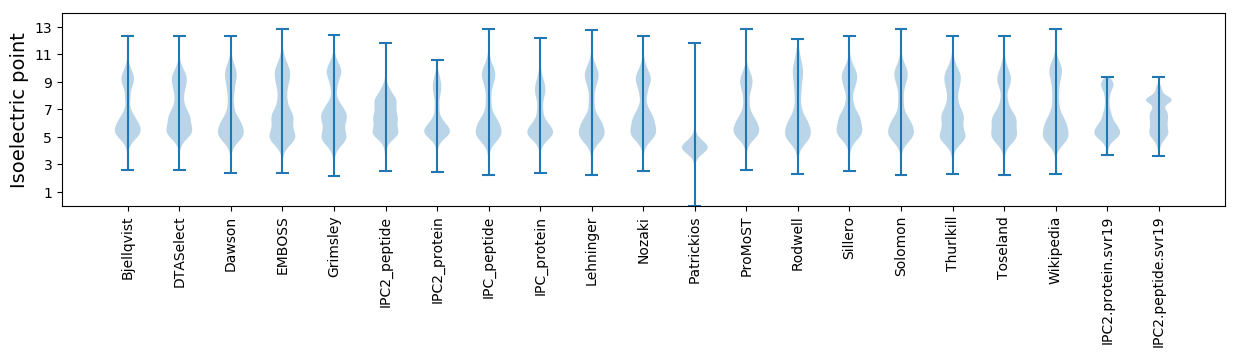
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K8QX80|A0A0K8QX80_9BACT Uncharacterized protein OS=Bacteroidales bacterium 6E OX=1688776 GN=BA6E_1093 PE=4 SV=1
MM1 pKa = 7.27YY2 pKa = 10.26ISLPLYY8 pKa = 9.01NDD10 pKa = 3.03NFIFCFSALINFNTASKK27 pKa = 10.53SSSKK31 pKa = 10.76KK32 pKa = 10.23FDD34 pKa = 3.34ISPLGCTRR42 pKa = 11.84KK43 pKa = 10.02NDD45 pKa = 3.68VLIGASFFLDD55 pKa = 4.54DD56 pKa = 3.65ILSIEE61 pKa = 4.2EE62 pKa = 4.37CNEE65 pKa = 3.34GCHH68 pKa = 6.58PFFCPRR74 pKa = 11.84ITPVVHH80 pKa = 6.42HH81 pKa = 6.92RR82 pKa = 11.84LHH84 pKa = 6.26GLVFVVACRR93 pKa = 11.84QPRR96 pKa = 11.84KK97 pKa = 9.82RR98 pKa = 11.84GGGVMRR104 pKa = 11.84RR105 pKa = 11.84CYY107 pKa = 10.6KK108 pKa = 10.21RR109 pKa = 11.84VSRR112 pKa = 11.84SLARR116 pKa = 11.84PGSKK120 pKa = 10.08RR121 pKa = 11.84IPTGLASLSDD131 pKa = 3.55PTRR134 pKa = 11.84CVGYY138 pKa = 10.38KK139 pKa = 9.88RR140 pKa = 11.84VIPTGLVFILIQRR153 pKa = 11.84IVLATNVSPARR164 pKa = 11.84WRR166 pKa = 11.84VRR168 pKa = 11.84DD169 pKa = 3.75RR170 pKa = 11.84NEE172 pKa = 4.36SQWDD176 pKa = 3.43WLRR179 pKa = 11.84SLIQRR184 pKa = 11.84VALVTNEE191 pKa = 3.34PPARR195 pKa = 11.84HH196 pKa = 6.41RR197 pKa = 11.84YY198 pKa = 9.17LSGRR202 pKa = 11.84PGSKK206 pKa = 9.3TNPDD210 pKa = 3.28GVQVGSLRR218 pKa = 3.62
MM1 pKa = 7.27YY2 pKa = 10.26ISLPLYY8 pKa = 9.01NDD10 pKa = 3.03NFIFCFSALINFNTASKK27 pKa = 10.53SSSKK31 pKa = 10.76KK32 pKa = 10.23FDD34 pKa = 3.34ISPLGCTRR42 pKa = 11.84KK43 pKa = 10.02NDD45 pKa = 3.68VLIGASFFLDD55 pKa = 4.54DD56 pKa = 3.65ILSIEE61 pKa = 4.2EE62 pKa = 4.37CNEE65 pKa = 3.34GCHH68 pKa = 6.58PFFCPRR74 pKa = 11.84ITPVVHH80 pKa = 6.42HH81 pKa = 6.92RR82 pKa = 11.84LHH84 pKa = 6.26GLVFVVACRR93 pKa = 11.84QPRR96 pKa = 11.84KK97 pKa = 9.82RR98 pKa = 11.84GGGVMRR104 pKa = 11.84RR105 pKa = 11.84CYY107 pKa = 10.6KK108 pKa = 10.21RR109 pKa = 11.84VSRR112 pKa = 11.84SLARR116 pKa = 11.84PGSKK120 pKa = 10.08RR121 pKa = 11.84IPTGLASLSDD131 pKa = 3.55PTRR134 pKa = 11.84CVGYY138 pKa = 10.38KK139 pKa = 9.88RR140 pKa = 11.84VIPTGLVFILIQRR153 pKa = 11.84IVLATNVSPARR164 pKa = 11.84WRR166 pKa = 11.84VRR168 pKa = 11.84DD169 pKa = 3.75RR170 pKa = 11.84NEE172 pKa = 4.36SQWDD176 pKa = 3.43WLRR179 pKa = 11.84SLIQRR184 pKa = 11.84VALVTNEE191 pKa = 3.34PPARR195 pKa = 11.84HH196 pKa = 6.41RR197 pKa = 11.84YY198 pKa = 9.17LSGRR202 pKa = 11.84PGSKK206 pKa = 9.3TNPDD210 pKa = 3.28GVQVGSLRR218 pKa = 3.62
Molecular weight: 24.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1318582 |
10 |
5663 |
364.5 |
41.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.755 ± 0.035 | 0.797 ± 0.019 |
5.509 ± 0.034 | 6.435 ± 0.04 |
4.934 ± 0.027 | 7.216 ± 0.043 |
2.014 ± 0.021 | 7.263 ± 0.034 |
5.866 ± 0.038 | 9.216 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.696 ± 0.022 | 5.113 ± 0.033 |
4.168 ± 0.023 | 3.412 ± 0.021 |
4.749 ± 0.036 | 6.413 ± 0.033 |
5.563 ± 0.059 | 6.603 ± 0.036 |
1.37 ± 0.017 | 3.907 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
