
Odonata-associated circular virus-1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
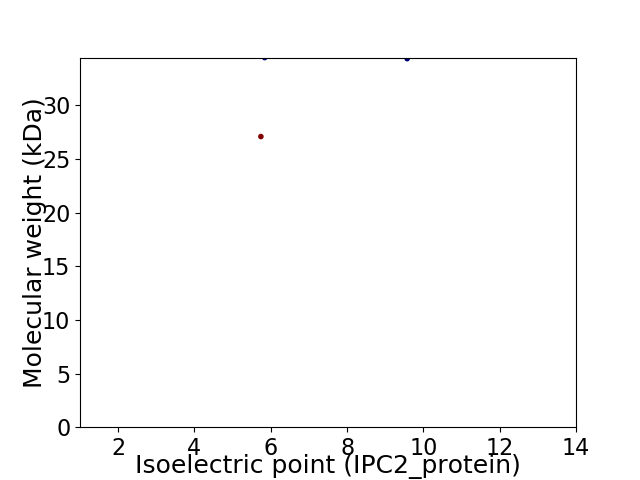
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0B4UGI4|A0A0B4UGI4_9VIRU Putative capsid protein OS=Odonata-associated circular virus-1 OX=1592109 PE=4 SV=1
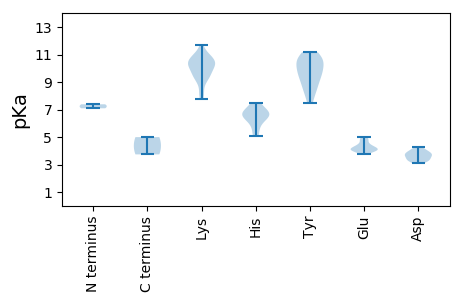
MM1 pKa = 7.38SAEE4 pKa = 4.01QPKK7 pKa = 10.4PSDD10 pKa = 3.81KK11 pKa = 10.79ASHH14 pKa = 6.58WLLTLNNPTADD25 pKa = 3.52DD26 pKa = 4.06RR27 pKa = 11.84EE28 pKa = 4.31RR29 pKa = 11.84CKK31 pKa = 10.78NLPKK35 pKa = 9.44WVKK38 pKa = 9.77RR39 pKa = 11.84FKK41 pKa = 10.77FQDD44 pKa = 3.68EE45 pKa = 4.23IGEE48 pKa = 4.38SGTLHH53 pKa = 5.11IQGYY57 pKa = 8.48IHH59 pKa = 6.94TEE61 pKa = 4.1TVRR64 pKa = 11.84LSALKK69 pKa = 9.34KK70 pKa = 8.18WFPRR74 pKa = 11.84GHH76 pKa = 6.48FEE78 pKa = 4.11VCRR81 pKa = 11.84NPKK84 pKa = 9.82AAEE87 pKa = 3.91NYY89 pKa = 9.43AGKK92 pKa = 10.51EE93 pKa = 4.06EE94 pKa = 4.26TSVPGTQHH102 pKa = 6.84DD103 pKa = 4.08FKK105 pKa = 11.13NTVEE109 pKa = 4.28QVTMMDD115 pKa = 3.52NLIRR119 pKa = 11.84LCDD122 pKa = 3.86IQPKK126 pKa = 10.27IIDD129 pKa = 3.32HH130 pKa = 7.5LIRR133 pKa = 11.84QNAVSTDD140 pKa = 3.1AKK142 pKa = 10.62QVLTGKK148 pKa = 9.51QLYY151 pKa = 8.51QEE153 pKa = 4.89EE154 pKa = 4.88YY155 pKa = 8.01WGCVNVILEE164 pKa = 4.59EE165 pKa = 4.24NPDD168 pKa = 4.27LISTYY173 pKa = 9.64TMPQYY178 pKa = 10.84CKK180 pKa = 10.26AWVNCRR186 pKa = 11.84PAILNAHH193 pKa = 6.63RR194 pKa = 11.84VRR196 pKa = 11.84QYY198 pKa = 10.82EE199 pKa = 3.97NEE201 pKa = 4.12RR202 pKa = 11.84TAGQPDD208 pKa = 3.98KK209 pKa = 11.71NEE211 pKa = 4.46IITPEE216 pKa = 4.11DD217 pKa = 3.43LTTNEE222 pKa = 4.79CLICSKK228 pKa = 10.36EE229 pKa = 4.02KK230 pKa = 11.03CEE232 pKa = 4.8CEE234 pKa = 3.81AA235 pKa = 4.99
MM1 pKa = 7.38SAEE4 pKa = 4.01QPKK7 pKa = 10.4PSDD10 pKa = 3.81KK11 pKa = 10.79ASHH14 pKa = 6.58WLLTLNNPTADD25 pKa = 3.52DD26 pKa = 4.06RR27 pKa = 11.84EE28 pKa = 4.31RR29 pKa = 11.84CKK31 pKa = 10.78NLPKK35 pKa = 9.44WVKK38 pKa = 9.77RR39 pKa = 11.84FKK41 pKa = 10.77FQDD44 pKa = 3.68EE45 pKa = 4.23IGEE48 pKa = 4.38SGTLHH53 pKa = 5.11IQGYY57 pKa = 8.48IHH59 pKa = 6.94TEE61 pKa = 4.1TVRR64 pKa = 11.84LSALKK69 pKa = 9.34KK70 pKa = 8.18WFPRR74 pKa = 11.84GHH76 pKa = 6.48FEE78 pKa = 4.11VCRR81 pKa = 11.84NPKK84 pKa = 9.82AAEE87 pKa = 3.91NYY89 pKa = 9.43AGKK92 pKa = 10.51EE93 pKa = 4.06EE94 pKa = 4.26TSVPGTQHH102 pKa = 6.84DD103 pKa = 4.08FKK105 pKa = 11.13NTVEE109 pKa = 4.28QVTMMDD115 pKa = 3.52NLIRR119 pKa = 11.84LCDD122 pKa = 3.86IQPKK126 pKa = 10.27IIDD129 pKa = 3.32HH130 pKa = 7.5LIRR133 pKa = 11.84QNAVSTDD140 pKa = 3.1AKK142 pKa = 10.62QVLTGKK148 pKa = 9.51QLYY151 pKa = 8.51QEE153 pKa = 4.89EE154 pKa = 4.88YY155 pKa = 8.01WGCVNVILEE164 pKa = 4.59EE165 pKa = 4.24NPDD168 pKa = 4.27LISTYY173 pKa = 9.64TMPQYY178 pKa = 10.84CKK180 pKa = 10.26AWVNCRR186 pKa = 11.84PAILNAHH193 pKa = 6.63RR194 pKa = 11.84VRR196 pKa = 11.84QYY198 pKa = 10.82EE199 pKa = 3.97NEE201 pKa = 4.12RR202 pKa = 11.84TAGQPDD208 pKa = 3.98KK209 pKa = 11.71NEE211 pKa = 4.46IITPEE216 pKa = 4.11DD217 pKa = 3.43LTTNEE222 pKa = 4.79CLICSKK228 pKa = 10.36EE229 pKa = 4.02KK230 pKa = 11.03CEE232 pKa = 4.8CEE234 pKa = 3.81AA235 pKa = 4.99
Molecular weight: 27.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGI4|A0A0B4UGI4_9VIRU Putative capsid protein OS=Odonata-associated circular virus-1 OX=1592109 PE=4 SV=1
MM1 pKa = 7.09VKK3 pKa = 10.21LVRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84VLRR12 pKa = 11.84KK13 pKa = 9.33RR14 pKa = 11.84KK15 pKa = 9.07SVPKK19 pKa = 9.47RR20 pKa = 11.84RR21 pKa = 11.84MMRR24 pKa = 11.84RR25 pKa = 11.84GVGRR29 pKa = 11.84ASVKK33 pKa = 9.56RR34 pKa = 11.84TSDD37 pKa = 3.29YY38 pKa = 11.17AKK40 pKa = 10.2CVEE43 pKa = 4.04IQEE46 pKa = 4.58TKK48 pKa = 7.78MTAVNDD54 pKa = 3.72ATLDD58 pKa = 3.74SVGGVINFCLQDD70 pKa = 3.24FQRR73 pKa = 11.84PQEE76 pKa = 3.76IAHH79 pKa = 6.53AYY81 pKa = 8.94KK82 pKa = 10.43YY83 pKa = 10.69YY84 pKa = 10.56RR85 pKa = 11.84AAKK88 pKa = 10.24CEE90 pKa = 3.96ITFIPYY96 pKa = 10.09FNIAQTANAAATQLPQLYY114 pKa = 7.46MTVDD118 pKa = 3.28RR119 pKa = 11.84LSNRR123 pKa = 11.84WIAPTEE129 pKa = 4.0SEE131 pKa = 4.09MLSRR135 pKa = 11.84GVSPRR140 pKa = 11.84LFTKK144 pKa = 10.43KK145 pKa = 8.99MRR147 pKa = 11.84LSFKK151 pKa = 10.64PNLLQGISLEE161 pKa = 4.44TQQSVNYY168 pKa = 9.51PSGSPPGGNPAGISNLGYY186 pKa = 10.09QKK188 pKa = 10.96AIAVFNKK195 pKa = 9.98GLPTQQSYY203 pKa = 10.72GYY205 pKa = 9.73SNNSTTGALQAGQVLAPLGVNPYY228 pKa = 10.24DD229 pKa = 3.6IRR231 pKa = 11.84YY232 pKa = 8.84HH233 pKa = 5.83GAAFVPAIEE242 pKa = 4.22VLAANTPVAVGDD254 pKa = 3.81IQVKK258 pKa = 8.22ITWEE262 pKa = 4.19SKK264 pKa = 9.99RR265 pKa = 11.84PRR267 pKa = 11.84ALMTNSPLNEE277 pKa = 4.99LIPYY281 pKa = 9.52VSSCCQGNTNAVLNTQPTSYY301 pKa = 10.55PSGAIGAAMTVLL313 pKa = 3.74
MM1 pKa = 7.09VKK3 pKa = 10.21LVRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84VLRR12 pKa = 11.84KK13 pKa = 9.33RR14 pKa = 11.84KK15 pKa = 9.07SVPKK19 pKa = 9.47RR20 pKa = 11.84RR21 pKa = 11.84MMRR24 pKa = 11.84RR25 pKa = 11.84GVGRR29 pKa = 11.84ASVKK33 pKa = 9.56RR34 pKa = 11.84TSDD37 pKa = 3.29YY38 pKa = 11.17AKK40 pKa = 10.2CVEE43 pKa = 4.04IQEE46 pKa = 4.58TKK48 pKa = 7.78MTAVNDD54 pKa = 3.72ATLDD58 pKa = 3.74SVGGVINFCLQDD70 pKa = 3.24FQRR73 pKa = 11.84PQEE76 pKa = 3.76IAHH79 pKa = 6.53AYY81 pKa = 8.94KK82 pKa = 10.43YY83 pKa = 10.69YY84 pKa = 10.56RR85 pKa = 11.84AAKK88 pKa = 10.24CEE90 pKa = 3.96ITFIPYY96 pKa = 10.09FNIAQTANAAATQLPQLYY114 pKa = 7.46MTVDD118 pKa = 3.28RR119 pKa = 11.84LSNRR123 pKa = 11.84WIAPTEE129 pKa = 4.0SEE131 pKa = 4.09MLSRR135 pKa = 11.84GVSPRR140 pKa = 11.84LFTKK144 pKa = 10.43KK145 pKa = 8.99MRR147 pKa = 11.84LSFKK151 pKa = 10.64PNLLQGISLEE161 pKa = 4.44TQQSVNYY168 pKa = 9.51PSGSPPGGNPAGISNLGYY186 pKa = 10.09QKK188 pKa = 10.96AIAVFNKK195 pKa = 9.98GLPTQQSYY203 pKa = 10.72GYY205 pKa = 9.73SNNSTTGALQAGQVLAPLGVNPYY228 pKa = 10.24DD229 pKa = 3.6IRR231 pKa = 11.84YY232 pKa = 8.84HH233 pKa = 5.83GAAFVPAIEE242 pKa = 4.22VLAANTPVAVGDD254 pKa = 3.81IQVKK258 pKa = 8.22ITWEE262 pKa = 4.19SKK264 pKa = 9.99RR265 pKa = 11.84PRR267 pKa = 11.84ALMTNSPLNEE277 pKa = 4.99LIPYY281 pKa = 9.52VSSCCQGNTNAVLNTQPTSYY301 pKa = 10.55PSGAIGAAMTVLL313 pKa = 3.74
Molecular weight: 34.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
548 |
235 |
313 |
274.0 |
30.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.212 ± 1.448 | 2.737 ± 0.975 |
3.467 ± 1.053 | 5.839 ± 2.262 |
2.372 ± 0.157 | 5.474 ± 1.056 |
1.642 ± 0.858 | 5.474 ± 0.31 |
6.204 ± 0.935 | 7.482 ± 0.159 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.43 | 6.204 ± 0.115 |
6.204 ± 0.432 | 5.657 ± 0.08 |
6.204 ± 0.705 | 5.839 ± 1.291 |
6.934 ± 0.192 | 6.569 ± 0.94 |
1.277 ± 0.546 | 3.832 ± 0.548 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
