
Pseudoalteromonas atlantica (strain T6c / ATCC BAA-1087)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas; Pseudoalteromonas atlantica
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
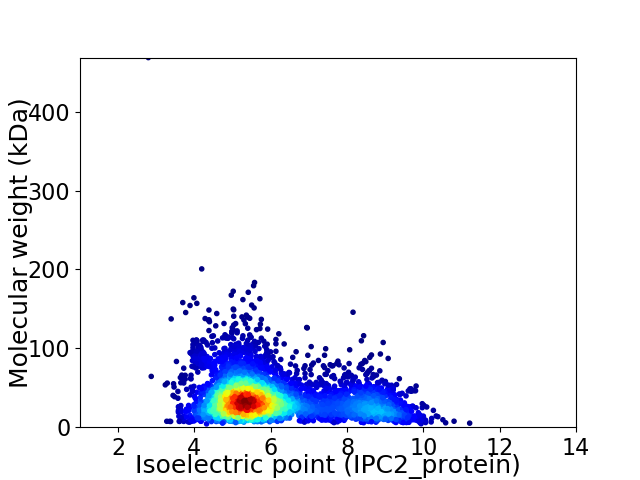
Virtual 2D-PAGE plot for 4271 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q15X31|RRAB_PSEA6 Regulator of ribonuclease activity B OS=Pseudoalteromonas atlantica (strain T6c / ATCC BAA-1087) OX=342610 GN=rraB PE=3 SV=1
MM1 pKa = 7.32KK2 pKa = 10.35LKK4 pKa = 10.88YY5 pKa = 9.28PALLALLSASAVQAGEE21 pKa = 4.08LVISGVFDD29 pKa = 3.98GPLPGGLPKK38 pKa = 10.7GIEE41 pKa = 4.03LFVTQDD47 pKa = 2.88IADD50 pKa = 4.47LSTCGVGSANNGGGTDD66 pKa = 3.44NQEE69 pKa = 3.78FTFPARR75 pKa = 11.84AATAGSYY82 pKa = 9.91IYY84 pKa = 10.25LASEE88 pKa = 4.53SPQFAAFFGFEE99 pKa = 3.84PTYY102 pKa = 10.5TNSAANINGDD112 pKa = 3.61DD113 pKa = 4.74AIEE116 pKa = 4.18LFCDD120 pKa = 4.02GAVVDD125 pKa = 4.5VFGDD129 pKa = 3.53INVDD133 pKa = 3.45GNGEE137 pKa = 3.76PWEE140 pKa = 4.14YY141 pKa = 11.13LDD143 pKa = 3.08GWAYY147 pKa = 11.15RR148 pKa = 11.84NIDD151 pKa = 3.59TGPDD155 pKa = 3.39GNSFLLGNWTFSGPNAFDD173 pKa = 4.03GEE175 pKa = 4.97SNNATAAAPFPLKK188 pKa = 10.83AFADD192 pKa = 4.78EE193 pKa = 4.43IDD195 pKa = 4.07GGDD198 pKa = 3.34TGGGSGGATDD208 pKa = 4.79PSAPCFNCPALDD220 pKa = 4.82PIADD224 pKa = 3.43ASTFDD229 pKa = 3.66SEE231 pKa = 4.61SYY233 pKa = 9.54YY234 pKa = 11.11AAVSAEE240 pKa = 3.85INANSPAATIKK251 pKa = 10.72SAITSAITQGQKK263 pKa = 10.33NLTYY267 pKa = 10.55SEE269 pKa = 4.1VWSALTHH276 pKa = 6.13TDD278 pKa = 3.66EE279 pKa = 6.2DD280 pKa = 4.64PNNPDD285 pKa = 5.86NVILWYY291 pKa = 10.46KK292 pKa = 8.02GTSIAKK298 pKa = 9.52SSNGSGSQSSDD309 pKa = 2.93PDD311 pKa = 2.92NWNRR315 pKa = 11.84EE316 pKa = 4.06HH317 pKa = 6.88SWAKK321 pKa = 10.09SHH323 pKa = 6.31GFPNEE328 pKa = 3.86SQEE331 pKa = 4.13AYY333 pKa = 9.83TDD335 pKa = 3.31IHH337 pKa = 6.53HH338 pKa = 7.15LRR340 pKa = 11.84PTDD343 pKa = 3.14ISVNSSRR350 pKa = 11.84GNLDD354 pKa = 3.51FDD356 pKa = 4.23NSDD359 pKa = 3.55NALSEE364 pKa = 4.43APSNRR369 pKa = 11.84VDD371 pKa = 4.12DD372 pKa = 5.33DD373 pKa = 3.96SFEE376 pKa = 4.31PRR378 pKa = 11.84DD379 pKa = 3.65AVKK382 pKa = 10.97GDD384 pKa = 3.46VARR387 pKa = 11.84MMFYY391 pKa = 9.97MDD393 pKa = 2.94TRR395 pKa = 11.84YY396 pKa = 10.6EE397 pKa = 4.2GFGDD401 pKa = 3.82STPDD405 pKa = 3.14LVLVDD410 pKa = 4.65RR411 pKa = 11.84LTSTGAPEE419 pKa = 4.55LGRR422 pKa = 11.84LCRR425 pKa = 11.84LVEE428 pKa = 3.79WHH430 pKa = 6.58NADD433 pKa = 3.77PVDD436 pKa = 3.59ATEE439 pKa = 3.82QTRR442 pKa = 11.84NDD444 pKa = 3.78TIYY447 pKa = 10.54EE448 pKa = 4.13YY449 pKa = 10.72QGNRR453 pKa = 11.84NPFIDD458 pKa = 3.68HH459 pKa = 7.07PEE461 pKa = 3.97WVDD464 pKa = 3.49ILYY467 pKa = 7.51QTPACGTDD475 pKa = 3.09GGDD478 pKa = 3.77TGGEE482 pKa = 4.07DD483 pKa = 5.01DD484 pKa = 5.38GDD486 pKa = 3.93TGGDD490 pKa = 3.32TGGSASAGDD499 pKa = 3.73IVISGVIDD507 pKa = 4.12GPLSGGTPKK516 pKa = 10.62AIEE519 pKa = 3.98FFVRR523 pKa = 11.84NDD525 pKa = 2.97IADD528 pKa = 4.13LSTCGFGSANNGGGSDD544 pKa = 3.51GQEE547 pKa = 3.85YY548 pKa = 8.62TFSGSAAAGDD558 pKa = 4.43FIYY561 pKa = 10.4IASEE565 pKa = 3.98GSNFDD570 pKa = 3.38AYY572 pKa = 10.91FGFSPTDD579 pKa = 3.16TAGAAGINGDD589 pKa = 3.82DD590 pKa = 4.97AIEE593 pKa = 4.21LFCDD597 pKa = 3.57GDD599 pKa = 4.04VVDD602 pKa = 5.34VFGDD606 pKa = 3.37INTDD610 pKa = 3.16GTGEE614 pKa = 4.08PWDD617 pKa = 4.75HH618 pKa = 6.26VDD620 pKa = 2.78GWAYY624 pKa = 10.17RR625 pKa = 11.84VASTGPDD632 pKa = 2.96GSTFTLSNWVFSGANALDD650 pKa = 4.07GEE652 pKa = 4.64ATNATAASQFPIGSYY667 pKa = 7.52TSKK670 pKa = 10.76EE671 pKa = 3.86VLLISGVFDD680 pKa = 4.25GPLSGGTPKK689 pKa = 10.6AIEE692 pKa = 4.7FYY694 pKa = 10.99AVTDD698 pKa = 3.69IQDD701 pKa = 3.42LSVFGFGSANNGGGTDD717 pKa = 3.58GQEE720 pKa = 4.09FAFSGSVSAGEE731 pKa = 4.05YY732 pKa = 10.15FYY734 pKa = 11.56VATEE738 pKa = 3.94TSGFNAYY745 pKa = 10.3FGFDD749 pKa = 3.25PTFISGAAAINGDD762 pKa = 3.69DD763 pKa = 4.52AIEE766 pKa = 4.31LFHH769 pKa = 7.16EE770 pKa = 5.06GEE772 pKa = 4.45VVDD775 pKa = 4.5VFGDD779 pKa = 3.58INVDD783 pKa = 3.36GTGQPWDD790 pKa = 4.16YY791 pKa = 11.39LDD793 pKa = 3.06GWAYY797 pKa = 10.18RR798 pKa = 11.84VADD801 pKa = 4.23SGPDD805 pKa = 3.04GTTFVLANWTFSGTNVLDD823 pKa = 4.21GSSNNATVSTPFPLGSFAGSGNNDD847 pKa = 3.14GGDD850 pKa = 3.52NGGGEE855 pKa = 4.35TPLLGMCADD864 pKa = 4.23PATLISAIQGSTDD877 pKa = 2.7ISPAVGEE884 pKa = 3.94THH886 pKa = 6.86IIEE889 pKa = 5.31GIVTASFPNLSGYY902 pKa = 9.38FVQEE906 pKa = 3.87EE907 pKa = 4.86DD908 pKa = 3.53SDD910 pKa = 3.95QDD912 pKa = 3.4ADD914 pKa = 3.76SATSEE919 pKa = 4.42GVFVYY924 pKa = 10.37SPSLALPEE932 pKa = 4.24AGKK935 pKa = 9.9VVRR938 pKa = 11.84VLGDD942 pKa = 3.1VAEE945 pKa = 4.89AFNKK949 pKa = 7.83TQLVVSEE956 pKa = 4.75INPDD960 pKa = 3.59CGTGSVTATALTLPFDD976 pKa = 4.38SVDD979 pKa = 3.36SMEE982 pKa = 4.72ALEE985 pKa = 4.5GMLVASSGEE994 pKa = 3.96LTVTDD999 pKa = 4.4TYY1001 pKa = 11.8SLGRR1005 pKa = 11.84YY1006 pKa = 9.83GEE1008 pKa = 4.31VLLSNGRR1015 pKa = 11.84LFIPTNLYY1023 pKa = 8.75TAGSPEE1029 pKa = 4.73AIALAAQNEE1038 pKa = 4.46LNQITLDD1045 pKa = 4.43DD1046 pKa = 4.51GVNGQNPDD1054 pKa = 3.39VVVYY1058 pKa = 8.7PAGGLSASNPLRR1070 pKa = 11.84GGDD1073 pKa = 3.44TVTALTGVMDD1083 pKa = 4.83FSFSLYY1089 pKa = 10.21RR1090 pKa = 11.84IIPVEE1095 pKa = 3.86QPTIVASNPRR1105 pKa = 11.84TDD1107 pKa = 4.89APDD1110 pKa = 3.99LEE1112 pKa = 4.89LGNLKK1117 pKa = 10.16VASLNVLNYY1126 pKa = 10.12FNTLDD1131 pKa = 3.81VSPNVCGPSNLEE1143 pKa = 3.7CRR1145 pKa = 11.84GADD1148 pKa = 3.55SEE1150 pKa = 5.05IEE1152 pKa = 4.28LEE1154 pKa = 4.12RR1155 pKa = 11.84QRR1157 pKa = 11.84AKK1159 pKa = 10.12TLAALLAMDD1168 pKa = 4.99ADD1170 pKa = 3.77IVGLMEE1176 pKa = 4.16VEE1178 pKa = 4.22NNGFGANSAVGDD1190 pKa = 4.15LVSGLNDD1197 pKa = 2.99VLGANTYY1204 pKa = 8.85ATVDD1208 pKa = 3.18AGSAIGTDD1216 pKa = 4.2AITVALIYY1224 pKa = 10.36KK1225 pKa = 8.28PAVVSVVGQPAILDD1239 pKa = 3.79SSNSISDD1246 pKa = 3.56DD1247 pKa = 4.06DD1248 pKa = 4.92GPLFLDD1254 pKa = 4.02TKK1256 pKa = 10.83NRR1258 pKa = 11.84PALNQKK1264 pKa = 9.66FALVANNEE1272 pKa = 4.11EE1273 pKa = 4.23LVVSVNHH1280 pKa = 6.38FKK1282 pKa = 11.2SKK1284 pKa = 10.55GSSCGAGDD1292 pKa = 5.25DD1293 pKa = 4.7DD1294 pKa = 4.38TTTGQGNCNLTRR1306 pKa = 11.84TRR1308 pKa = 11.84AAEE1311 pKa = 3.93ALTTFLSQQFSDD1323 pKa = 3.9TPTLIIGDD1331 pKa = 4.12LNSYY1335 pKa = 10.95AMEE1338 pKa = 4.17EE1339 pKa = 4.48PILKK1343 pKa = 9.73ILQQGYY1349 pKa = 7.98TDD1351 pKa = 4.4LANKK1355 pKa = 10.27FGGDD1359 pKa = 3.12EE1360 pKa = 4.27AYY1362 pKa = 10.29SYY1364 pKa = 11.73SFGGEE1369 pKa = 3.82FGYY1372 pKa = 10.74LDD1374 pKa = 3.95HH1375 pKa = 7.3ALASATLVDD1384 pKa = 4.24QIVDD1388 pKa = 3.5TTEE1391 pKa = 3.09WHH1393 pKa = 6.54INADD1397 pKa = 3.63EE1398 pKa = 5.23PIVFDD1403 pKa = 4.06YY1404 pKa = 11.16NVEE1407 pKa = 4.33FKK1409 pKa = 10.85SDD1411 pKa = 3.4QQLIDD1416 pKa = 3.76YY1417 pKa = 7.18YY1418 pKa = 11.58APDD1421 pKa = 4.21AYY1423 pKa = 10.77RR1424 pKa = 11.84MSDD1427 pKa = 3.35HH1428 pKa = 7.17DD1429 pKa = 4.07PVVISLLLEE1438 pKa = 4.22SEE1440 pKa = 4.54VQVVEE1445 pKa = 4.0GDD1447 pKa = 3.35YY1448 pKa = 11.53DD1449 pKa = 4.23GDD1451 pKa = 4.05GDD1453 pKa = 4.35VDD1455 pKa = 3.75MMDD1458 pKa = 2.87IRR1460 pKa = 11.84ALTRR1464 pKa = 11.84AIQLRR1469 pKa = 11.84QTIDD1473 pKa = 2.93NSFDD1477 pKa = 3.65FNEE1480 pKa = 4.84DD1481 pKa = 3.21GQVSYY1486 pKa = 11.41TDD1488 pKa = 3.24VRR1490 pKa = 11.84LLQRR1494 pKa = 11.84MCTRR1498 pKa = 11.84TRR1500 pKa = 11.84CAII1503 pKa = 3.63
MM1 pKa = 7.32KK2 pKa = 10.35LKK4 pKa = 10.88YY5 pKa = 9.28PALLALLSASAVQAGEE21 pKa = 4.08LVISGVFDD29 pKa = 3.98GPLPGGLPKK38 pKa = 10.7GIEE41 pKa = 4.03LFVTQDD47 pKa = 2.88IADD50 pKa = 4.47LSTCGVGSANNGGGTDD66 pKa = 3.44NQEE69 pKa = 3.78FTFPARR75 pKa = 11.84AATAGSYY82 pKa = 9.91IYY84 pKa = 10.25LASEE88 pKa = 4.53SPQFAAFFGFEE99 pKa = 3.84PTYY102 pKa = 10.5TNSAANINGDD112 pKa = 3.61DD113 pKa = 4.74AIEE116 pKa = 4.18LFCDD120 pKa = 4.02GAVVDD125 pKa = 4.5VFGDD129 pKa = 3.53INVDD133 pKa = 3.45GNGEE137 pKa = 3.76PWEE140 pKa = 4.14YY141 pKa = 11.13LDD143 pKa = 3.08GWAYY147 pKa = 11.15RR148 pKa = 11.84NIDD151 pKa = 3.59TGPDD155 pKa = 3.39GNSFLLGNWTFSGPNAFDD173 pKa = 4.03GEE175 pKa = 4.97SNNATAAAPFPLKK188 pKa = 10.83AFADD192 pKa = 4.78EE193 pKa = 4.43IDD195 pKa = 4.07GGDD198 pKa = 3.34TGGGSGGATDD208 pKa = 4.79PSAPCFNCPALDD220 pKa = 4.82PIADD224 pKa = 3.43ASTFDD229 pKa = 3.66SEE231 pKa = 4.61SYY233 pKa = 9.54YY234 pKa = 11.11AAVSAEE240 pKa = 3.85INANSPAATIKK251 pKa = 10.72SAITSAITQGQKK263 pKa = 10.33NLTYY267 pKa = 10.55SEE269 pKa = 4.1VWSALTHH276 pKa = 6.13TDD278 pKa = 3.66EE279 pKa = 6.2DD280 pKa = 4.64PNNPDD285 pKa = 5.86NVILWYY291 pKa = 10.46KK292 pKa = 8.02GTSIAKK298 pKa = 9.52SSNGSGSQSSDD309 pKa = 2.93PDD311 pKa = 2.92NWNRR315 pKa = 11.84EE316 pKa = 4.06HH317 pKa = 6.88SWAKK321 pKa = 10.09SHH323 pKa = 6.31GFPNEE328 pKa = 3.86SQEE331 pKa = 4.13AYY333 pKa = 9.83TDD335 pKa = 3.31IHH337 pKa = 6.53HH338 pKa = 7.15LRR340 pKa = 11.84PTDD343 pKa = 3.14ISVNSSRR350 pKa = 11.84GNLDD354 pKa = 3.51FDD356 pKa = 4.23NSDD359 pKa = 3.55NALSEE364 pKa = 4.43APSNRR369 pKa = 11.84VDD371 pKa = 4.12DD372 pKa = 5.33DD373 pKa = 3.96SFEE376 pKa = 4.31PRR378 pKa = 11.84DD379 pKa = 3.65AVKK382 pKa = 10.97GDD384 pKa = 3.46VARR387 pKa = 11.84MMFYY391 pKa = 9.97MDD393 pKa = 2.94TRR395 pKa = 11.84YY396 pKa = 10.6EE397 pKa = 4.2GFGDD401 pKa = 3.82STPDD405 pKa = 3.14LVLVDD410 pKa = 4.65RR411 pKa = 11.84LTSTGAPEE419 pKa = 4.55LGRR422 pKa = 11.84LCRR425 pKa = 11.84LVEE428 pKa = 3.79WHH430 pKa = 6.58NADD433 pKa = 3.77PVDD436 pKa = 3.59ATEE439 pKa = 3.82QTRR442 pKa = 11.84NDD444 pKa = 3.78TIYY447 pKa = 10.54EE448 pKa = 4.13YY449 pKa = 10.72QGNRR453 pKa = 11.84NPFIDD458 pKa = 3.68HH459 pKa = 7.07PEE461 pKa = 3.97WVDD464 pKa = 3.49ILYY467 pKa = 7.51QTPACGTDD475 pKa = 3.09GGDD478 pKa = 3.77TGGEE482 pKa = 4.07DD483 pKa = 5.01DD484 pKa = 5.38GDD486 pKa = 3.93TGGDD490 pKa = 3.32TGGSASAGDD499 pKa = 3.73IVISGVIDD507 pKa = 4.12GPLSGGTPKK516 pKa = 10.62AIEE519 pKa = 3.98FFVRR523 pKa = 11.84NDD525 pKa = 2.97IADD528 pKa = 4.13LSTCGFGSANNGGGSDD544 pKa = 3.51GQEE547 pKa = 3.85YY548 pKa = 8.62TFSGSAAAGDD558 pKa = 4.43FIYY561 pKa = 10.4IASEE565 pKa = 3.98GSNFDD570 pKa = 3.38AYY572 pKa = 10.91FGFSPTDD579 pKa = 3.16TAGAAGINGDD589 pKa = 3.82DD590 pKa = 4.97AIEE593 pKa = 4.21LFCDD597 pKa = 3.57GDD599 pKa = 4.04VVDD602 pKa = 5.34VFGDD606 pKa = 3.37INTDD610 pKa = 3.16GTGEE614 pKa = 4.08PWDD617 pKa = 4.75HH618 pKa = 6.26VDD620 pKa = 2.78GWAYY624 pKa = 10.17RR625 pKa = 11.84VASTGPDD632 pKa = 2.96GSTFTLSNWVFSGANALDD650 pKa = 4.07GEE652 pKa = 4.64ATNATAASQFPIGSYY667 pKa = 7.52TSKK670 pKa = 10.76EE671 pKa = 3.86VLLISGVFDD680 pKa = 4.25GPLSGGTPKK689 pKa = 10.6AIEE692 pKa = 4.7FYY694 pKa = 10.99AVTDD698 pKa = 3.69IQDD701 pKa = 3.42LSVFGFGSANNGGGTDD717 pKa = 3.58GQEE720 pKa = 4.09FAFSGSVSAGEE731 pKa = 4.05YY732 pKa = 10.15FYY734 pKa = 11.56VATEE738 pKa = 3.94TSGFNAYY745 pKa = 10.3FGFDD749 pKa = 3.25PTFISGAAAINGDD762 pKa = 3.69DD763 pKa = 4.52AIEE766 pKa = 4.31LFHH769 pKa = 7.16EE770 pKa = 5.06GEE772 pKa = 4.45VVDD775 pKa = 4.5VFGDD779 pKa = 3.58INVDD783 pKa = 3.36GTGQPWDD790 pKa = 4.16YY791 pKa = 11.39LDD793 pKa = 3.06GWAYY797 pKa = 10.18RR798 pKa = 11.84VADD801 pKa = 4.23SGPDD805 pKa = 3.04GTTFVLANWTFSGTNVLDD823 pKa = 4.21GSSNNATVSTPFPLGSFAGSGNNDD847 pKa = 3.14GGDD850 pKa = 3.52NGGGEE855 pKa = 4.35TPLLGMCADD864 pKa = 4.23PATLISAIQGSTDD877 pKa = 2.7ISPAVGEE884 pKa = 3.94THH886 pKa = 6.86IIEE889 pKa = 5.31GIVTASFPNLSGYY902 pKa = 9.38FVQEE906 pKa = 3.87EE907 pKa = 4.86DD908 pKa = 3.53SDD910 pKa = 3.95QDD912 pKa = 3.4ADD914 pKa = 3.76SATSEE919 pKa = 4.42GVFVYY924 pKa = 10.37SPSLALPEE932 pKa = 4.24AGKK935 pKa = 9.9VVRR938 pKa = 11.84VLGDD942 pKa = 3.1VAEE945 pKa = 4.89AFNKK949 pKa = 7.83TQLVVSEE956 pKa = 4.75INPDD960 pKa = 3.59CGTGSVTATALTLPFDD976 pKa = 4.38SVDD979 pKa = 3.36SMEE982 pKa = 4.72ALEE985 pKa = 4.5GMLVASSGEE994 pKa = 3.96LTVTDD999 pKa = 4.4TYY1001 pKa = 11.8SLGRR1005 pKa = 11.84YY1006 pKa = 9.83GEE1008 pKa = 4.31VLLSNGRR1015 pKa = 11.84LFIPTNLYY1023 pKa = 8.75TAGSPEE1029 pKa = 4.73AIALAAQNEE1038 pKa = 4.46LNQITLDD1045 pKa = 4.43DD1046 pKa = 4.51GVNGQNPDD1054 pKa = 3.39VVVYY1058 pKa = 8.7PAGGLSASNPLRR1070 pKa = 11.84GGDD1073 pKa = 3.44TVTALTGVMDD1083 pKa = 4.83FSFSLYY1089 pKa = 10.21RR1090 pKa = 11.84IIPVEE1095 pKa = 3.86QPTIVASNPRR1105 pKa = 11.84TDD1107 pKa = 4.89APDD1110 pKa = 3.99LEE1112 pKa = 4.89LGNLKK1117 pKa = 10.16VASLNVLNYY1126 pKa = 10.12FNTLDD1131 pKa = 3.81VSPNVCGPSNLEE1143 pKa = 3.7CRR1145 pKa = 11.84GADD1148 pKa = 3.55SEE1150 pKa = 5.05IEE1152 pKa = 4.28LEE1154 pKa = 4.12RR1155 pKa = 11.84QRR1157 pKa = 11.84AKK1159 pKa = 10.12TLAALLAMDD1168 pKa = 4.99ADD1170 pKa = 3.77IVGLMEE1176 pKa = 4.16VEE1178 pKa = 4.22NNGFGANSAVGDD1190 pKa = 4.15LVSGLNDD1197 pKa = 2.99VLGANTYY1204 pKa = 8.85ATVDD1208 pKa = 3.18AGSAIGTDD1216 pKa = 4.2AITVALIYY1224 pKa = 10.36KK1225 pKa = 8.28PAVVSVVGQPAILDD1239 pKa = 3.79SSNSISDD1246 pKa = 3.56DD1247 pKa = 4.06DD1248 pKa = 4.92GPLFLDD1254 pKa = 4.02TKK1256 pKa = 10.83NRR1258 pKa = 11.84PALNQKK1264 pKa = 9.66FALVANNEE1272 pKa = 4.11EE1273 pKa = 4.23LVVSVNHH1280 pKa = 6.38FKK1282 pKa = 11.2SKK1284 pKa = 10.55GSSCGAGDD1292 pKa = 5.25DD1293 pKa = 4.7DD1294 pKa = 4.38TTTGQGNCNLTRR1306 pKa = 11.84TRR1308 pKa = 11.84AAEE1311 pKa = 3.93ALTTFLSQQFSDD1323 pKa = 3.9TPTLIIGDD1331 pKa = 4.12LNSYY1335 pKa = 10.95AMEE1338 pKa = 4.17EE1339 pKa = 4.48PILKK1343 pKa = 9.73ILQQGYY1349 pKa = 7.98TDD1351 pKa = 4.4LANKK1355 pKa = 10.27FGGDD1359 pKa = 3.12EE1360 pKa = 4.27AYY1362 pKa = 10.29SYY1364 pKa = 11.73SFGGEE1369 pKa = 3.82FGYY1372 pKa = 10.74LDD1374 pKa = 3.95HH1375 pKa = 7.3ALASATLVDD1384 pKa = 4.24QIVDD1388 pKa = 3.5TTEE1391 pKa = 3.09WHH1393 pKa = 6.54INADD1397 pKa = 3.63EE1398 pKa = 5.23PIVFDD1403 pKa = 4.06YY1404 pKa = 11.16NVEE1407 pKa = 4.33FKK1409 pKa = 10.85SDD1411 pKa = 3.4QQLIDD1416 pKa = 3.76YY1417 pKa = 7.18YY1418 pKa = 11.58APDD1421 pKa = 4.21AYY1423 pKa = 10.77RR1424 pKa = 11.84MSDD1427 pKa = 3.35HH1428 pKa = 7.17DD1429 pKa = 4.07PVVISLLLEE1438 pKa = 4.22SEE1440 pKa = 4.54VQVVEE1445 pKa = 4.0GDD1447 pKa = 3.35YY1448 pKa = 11.53DD1449 pKa = 4.23GDD1451 pKa = 4.05GDD1453 pKa = 4.35VDD1455 pKa = 3.75MMDD1458 pKa = 2.87IRR1460 pKa = 11.84ALTRR1464 pKa = 11.84AIQLRR1469 pKa = 11.84QTIDD1473 pKa = 2.93NSFDD1477 pKa = 3.65FNEE1480 pKa = 4.84DD1481 pKa = 3.21GQVSYY1486 pKa = 11.41TDD1488 pKa = 3.24VRR1490 pKa = 11.84LLQRR1494 pKa = 11.84MCTRR1498 pKa = 11.84TRR1500 pKa = 11.84CAII1503 pKa = 3.63
Molecular weight: 158.02 kDa
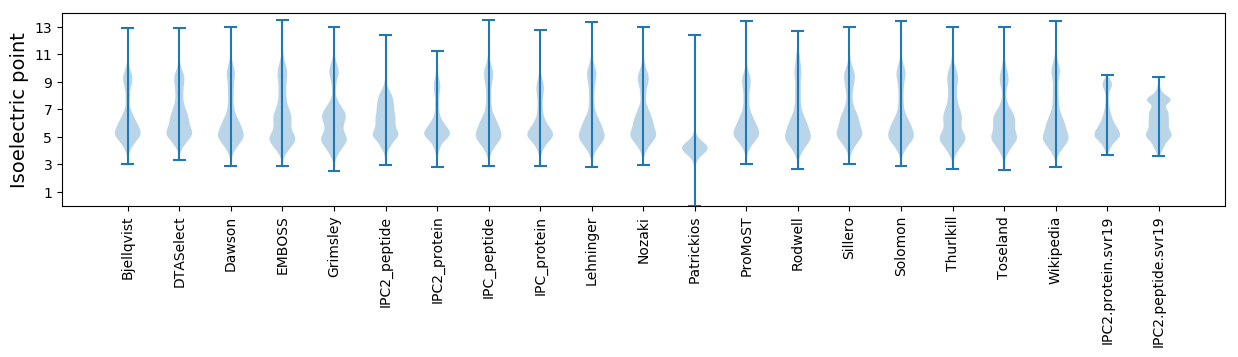
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q15MT0|Q15MT0_PSEA6 Flavodoxin/nitric oxide synthase OS=Pseudoalteromonas atlantica (strain T6c / ATCC BAA-1087) OX=342610 GN=Patl_4309 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.34GRR39 pKa = 11.84ARR41 pKa = 11.84LTAA44 pKa = 4.19
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.34GRR39 pKa = 11.84ARR41 pKa = 11.84LTAA44 pKa = 4.19
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1485885 |
33 |
4689 |
347.9 |
38.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.853 ± 0.035 | 0.981 ± 0.012 |
5.848 ± 0.032 | 5.786 ± 0.03 |
4.287 ± 0.025 | 6.82 ± 0.034 |
2.304 ± 0.023 | 6.189 ± 0.028 |
5.049 ± 0.037 | 10.114 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 0.021 | 4.499 ± 0.027 |
3.939 ± 0.02 | 4.775 ± 0.03 |
4.393 ± 0.029 | 6.912 ± 0.032 |
5.478 ± 0.05 | 6.913 ± 0.032 |
1.243 ± 0.015 | 3.108 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
