
Brachybacterium nesterenkovii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermabacteraceae; Brachybacterium
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
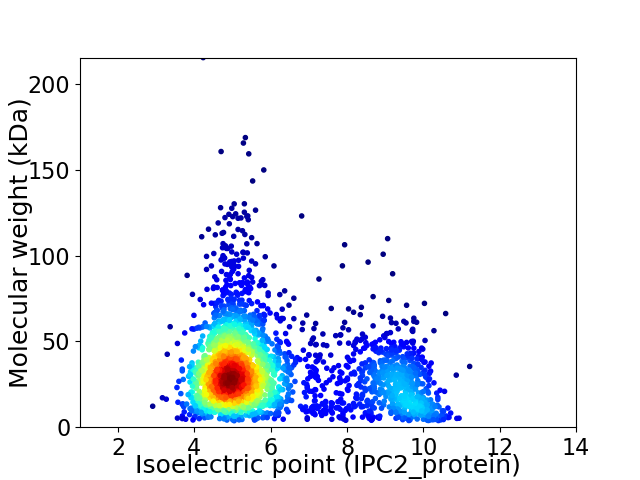
Virtual 2D-PAGE plot for 2675 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X6X8E5|A0A1X6X8E5_9MICO Cytidine deaminase OS=Brachybacterium nesterenkovii OX=47847 GN=FM110_12780 PE=4 SV=1
MM1 pKa = 7.7PGAAYY6 pKa = 7.85PATATGAIPAQPGAMPVHH24 pKa = 6.81PGGFGVPGAMPTMTGAIPAQQTGQLPTATGAIPVQPTAQLPTATGAIPAQSAGPAPSALAPHH86 pKa = 6.59EE87 pKa = 5.14LVAQPWPAVPGTTHH101 pKa = 7.02GSGAPRR107 pKa = 11.84SRR109 pKa = 11.84AGLVAAIAGLCALLLVLLAGLTAAGIYY136 pKa = 9.33VAASLQSMEE145 pKa = 4.2GTIEE149 pKa = 3.87QAPAPGTPGGDD160 pKa = 3.31EE161 pKa = 4.16PQTVPGTVLDD171 pKa = 3.94SSGAPVDD178 pKa = 4.2GLGTPDD184 pKa = 3.65APAIMGEE191 pKa = 4.04NSLRR195 pKa = 11.84WPTEE199 pKa = 3.78DD200 pKa = 4.79GGTLTLEE207 pKa = 4.39IEE209 pKa = 4.56SVTWNADD216 pKa = 3.08EE217 pKa = 5.64DD218 pKa = 4.2IKK220 pKa = 11.44ASDD223 pKa = 3.91PSAPAPEE230 pKa = 5.83AGMHH234 pKa = 5.84YY235 pKa = 11.09ALITIRR241 pKa = 11.84GTYY244 pKa = 9.52EE245 pKa = 3.71GEE247 pKa = 4.07KK248 pKa = 10.43YY249 pKa = 10.57AVLGDD254 pKa = 4.97DD255 pKa = 3.56IDD257 pKa = 4.48LAIEE261 pKa = 3.98TDD263 pKa = 3.01AGVYY267 pKa = 10.39YY268 pKa = 10.66DD269 pKa = 5.07DD270 pKa = 4.23GQIRR274 pKa = 11.84APEE277 pKa = 4.1PLWRR281 pKa = 11.84MGGITDD287 pKa = 4.27GEE289 pKa = 4.57SASGQMVIAVPEE301 pKa = 4.22AEE303 pKa = 4.27VGSALVSVAPWGGDD317 pKa = 3.24YY318 pKa = 10.95LYY320 pKa = 10.94VAEE323 pKa = 4.55SS324 pKa = 3.19
MM1 pKa = 7.7PGAAYY6 pKa = 7.85PATATGAIPAQPGAMPVHH24 pKa = 6.81PGGFGVPGAMPTMTGAIPAQQTGQLPTATGAIPVQPTAQLPTATGAIPAQSAGPAPSALAPHH86 pKa = 6.59EE87 pKa = 5.14LVAQPWPAVPGTTHH101 pKa = 7.02GSGAPRR107 pKa = 11.84SRR109 pKa = 11.84AGLVAAIAGLCALLLVLLAGLTAAGIYY136 pKa = 9.33VAASLQSMEE145 pKa = 4.2GTIEE149 pKa = 3.87QAPAPGTPGGDD160 pKa = 3.31EE161 pKa = 4.16PQTVPGTVLDD171 pKa = 3.94SSGAPVDD178 pKa = 4.2GLGTPDD184 pKa = 3.65APAIMGEE191 pKa = 4.04NSLRR195 pKa = 11.84WPTEE199 pKa = 3.78DD200 pKa = 4.79GGTLTLEE207 pKa = 4.39IEE209 pKa = 4.56SVTWNADD216 pKa = 3.08EE217 pKa = 5.64DD218 pKa = 4.2IKK220 pKa = 11.44ASDD223 pKa = 3.91PSAPAPEE230 pKa = 5.83AGMHH234 pKa = 5.84YY235 pKa = 11.09ALITIRR241 pKa = 11.84GTYY244 pKa = 9.52EE245 pKa = 3.71GEE247 pKa = 4.07KK248 pKa = 10.43YY249 pKa = 10.57AVLGDD254 pKa = 4.97DD255 pKa = 3.56IDD257 pKa = 4.48LAIEE261 pKa = 3.98TDD263 pKa = 3.01AGVYY267 pKa = 10.39YY268 pKa = 10.66DD269 pKa = 5.07DD270 pKa = 4.23GQIRR274 pKa = 11.84APEE277 pKa = 4.1PLWRR281 pKa = 11.84MGGITDD287 pKa = 4.27GEE289 pKa = 4.57SASGQMVIAVPEE301 pKa = 4.22AEE303 pKa = 4.27VGSALVSVAPWGGDD317 pKa = 3.24YY318 pKa = 10.95LYY320 pKa = 10.94VAEE323 pKa = 4.55SS324 pKa = 3.19
Molecular weight: 32.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X6X943|A0A1X6X943_9MICO Uncharacterized protein OS=Brachybacterium nesterenkovii OX=47847 GN=FM110_13165 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84TTLTTTHH9 pKa = 7.02RR10 pKa = 11.84AITMRR15 pKa = 11.84TTLTVTLGTVAIGAALAIALGTVTVGTLRR44 pKa = 11.84AVAIGALLAIALRR57 pKa = 11.84AVSIGPALSVTPRR70 pKa = 11.84SITVGPTLPTTLGAVTVGPTLPIALRR96 pKa = 11.84PIAIGATLAVTPRR109 pKa = 11.84PITVRR114 pKa = 11.84TTLALALGTVTVGTLRR130 pKa = 11.84AVAIGALLAIALRR143 pKa = 11.84PIPIRR148 pKa = 11.84TTLALALGAITVGPTLPVTLRR169 pKa = 11.84PITIRR174 pKa = 11.84TTLPIPLRR182 pKa = 11.84ATAVGPTLPVTLRR195 pKa = 11.84TITVRR200 pKa = 11.84TTLPIPLGTVTVGTLRR216 pKa = 11.84AVAIGATLAVTPRR229 pKa = 11.84PITVRR234 pKa = 11.84TALALALRR242 pKa = 11.84TITVGPTLPVTLRR255 pKa = 11.84PITVGPTLTITLGAITVRR273 pKa = 11.84TTLPIPLGAITVRR286 pKa = 11.84TTLPITLRR294 pKa = 11.84TIPVRR299 pKa = 11.84PTPTIVSRR307 pKa = 11.84RR308 pKa = 11.84AAAGAHH314 pKa = 6.37AARR317 pKa = 11.84ALVARR322 pKa = 11.84SVVVSVFHH330 pKa = 6.56DD331 pKa = 3.57QPLFASADD339 pKa = 3.47VRR341 pKa = 11.84AGAVVPPRR349 pKa = 3.73
MM1 pKa = 7.55RR2 pKa = 11.84TTLTTTHH9 pKa = 7.02RR10 pKa = 11.84AITMRR15 pKa = 11.84TTLTVTLGTVAIGAALAIALGTVTVGTLRR44 pKa = 11.84AVAIGALLAIALRR57 pKa = 11.84AVSIGPALSVTPRR70 pKa = 11.84SITVGPTLPTTLGAVTVGPTLPIALRR96 pKa = 11.84PIAIGATLAVTPRR109 pKa = 11.84PITVRR114 pKa = 11.84TTLALALGTVTVGTLRR130 pKa = 11.84AVAIGALLAIALRR143 pKa = 11.84PIPIRR148 pKa = 11.84TTLALALGAITVGPTLPVTLRR169 pKa = 11.84PITIRR174 pKa = 11.84TTLPIPLRR182 pKa = 11.84ATAVGPTLPVTLRR195 pKa = 11.84TITVRR200 pKa = 11.84TTLPIPLGTVTVGTLRR216 pKa = 11.84AVAIGATLAVTPRR229 pKa = 11.84PITVRR234 pKa = 11.84TALALALRR242 pKa = 11.84TITVGPTLPVTLRR255 pKa = 11.84PITVGPTLTITLGAITVRR273 pKa = 11.84TTLPIPLGAITVRR286 pKa = 11.84TTLPITLRR294 pKa = 11.84TIPVRR299 pKa = 11.84PTPTIVSRR307 pKa = 11.84RR308 pKa = 11.84AAAGAHH314 pKa = 6.37AARR317 pKa = 11.84ALVARR322 pKa = 11.84SVVVSVFHH330 pKa = 6.56DD331 pKa = 3.57QPLFASADD339 pKa = 3.47VRR341 pKa = 11.84AGAVVPPRR349 pKa = 3.73
Molecular weight: 35.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870157 |
37 |
2070 |
325.3 |
34.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.304 ± 0.073 | 0.651 ± 0.013 |
6.405 ± 0.043 | 6.029 ± 0.044 |
2.563 ± 0.031 | 9.242 ± 0.046 |
2.113 ± 0.02 | 4.168 ± 0.031 |
1.64 ± 0.037 | 10.16 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.022 | 1.445 ± 0.02 |
5.842 ± 0.036 | 2.671 ± 0.025 |
8.118 ± 0.058 | 5.402 ± 0.035 |
5.896 ± 0.036 | 8.343 ± 0.043 |
1.34 ± 0.018 | 1.663 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
