
Sewage-associated circular DNA virus-21
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
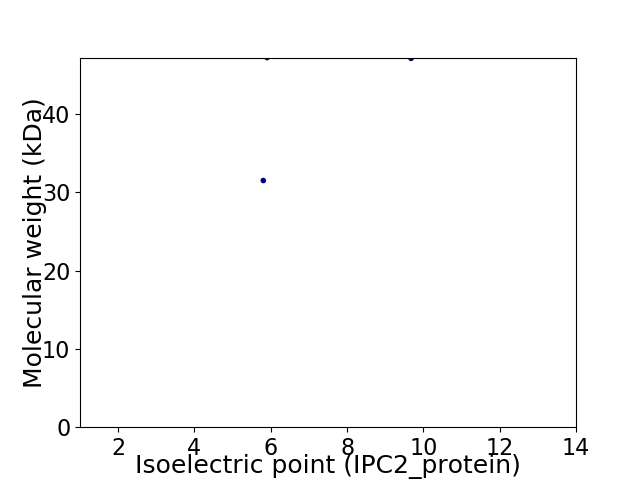
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0B4UI44|A0A0B4UI44_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-21 OX=1592088 PE=3 SV=1
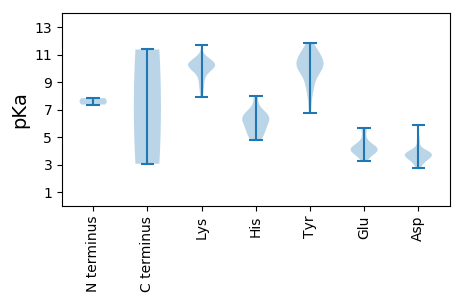
MM1 pKa = 7.83SSRR4 pKa = 11.84AWCFTLNNYY13 pKa = 9.94SEE15 pKa = 5.67DD16 pKa = 3.77DD17 pKa = 3.69LFEE20 pKa = 5.67ISGWDD25 pKa = 3.33CKK27 pKa = 10.6YY28 pKa = 10.38VVYY31 pKa = 10.14GVEE34 pKa = 4.13VAPEE38 pKa = 3.73TGTRR42 pKa = 11.84HH43 pKa = 4.76LQGYY47 pKa = 10.0CMFLGVQRR55 pKa = 11.84LSAVKK60 pKa = 10.24KK61 pKa = 10.43LLEE64 pKa = 4.09TAHH67 pKa = 6.32WEE69 pKa = 4.0IARR72 pKa = 11.84GSPLQNRR79 pKa = 11.84TYY81 pKa = 9.29CTKK84 pKa = 10.32EE85 pKa = 3.43GDD87 pKa = 3.78FVEE90 pKa = 5.21VGTLPLPRR98 pKa = 11.84GEE100 pKa = 4.29SEE102 pKa = 3.9KK103 pKa = 10.75EE104 pKa = 3.26RR105 pKa = 11.84WAEE108 pKa = 3.55AFEE111 pKa = 4.2AAKK114 pKa = 10.27HH115 pKa = 5.48GRR117 pKa = 11.84LDD119 pKa = 5.85DD120 pKa = 3.77IPEE123 pKa = 4.91DD124 pKa = 3.22IRR126 pKa = 11.84VRR128 pKa = 11.84YY129 pKa = 8.19YY130 pKa = 10.35RR131 pKa = 11.84TMKK134 pKa = 10.17EE135 pKa = 3.44IKK137 pKa = 8.78KK138 pKa = 9.97DD139 pKa = 3.59HH140 pKa = 5.62MVKK143 pKa = 10.6PEE145 pKa = 4.01DD146 pKa = 3.73QQDD149 pKa = 3.66VTGEE153 pKa = 3.9WFYY156 pKa = 11.85GEE158 pKa = 4.23PGVGKK163 pKa = 9.87SHH165 pKa = 7.95LARR168 pKa = 11.84EE169 pKa = 4.36QYY171 pKa = 10.04PGAYY175 pKa = 10.26LKK177 pKa = 9.28MQNKK181 pKa = 7.88WWDD184 pKa = 3.65GYY186 pKa = 10.21QDD188 pKa = 3.6EE189 pKa = 5.5KK190 pKa = 11.67YY191 pKa = 11.06VILDD195 pKa = 3.9DD196 pKa = 4.75FDD198 pKa = 5.07CKK200 pKa = 10.88EE201 pKa = 4.46LGHH204 pKa = 6.56HH205 pKa = 6.25LKK207 pKa = 9.94IWGDD211 pKa = 3.33KK212 pKa = 10.54YY213 pKa = 11.64SFLAEE218 pKa = 4.23TKK220 pKa = 10.47GGAICIRR227 pKa = 11.84PEE229 pKa = 3.84KK230 pKa = 10.78LIVTSNYY237 pKa = 9.74KK238 pKa = 10.2PEE240 pKa = 3.98QLFSDD245 pKa = 3.84EE246 pKa = 3.99MLIAVKK252 pKa = 10.35RR253 pKa = 11.84RR254 pKa = 11.84FKK256 pKa = 9.56VTHH259 pKa = 5.07VLSWRR264 pKa = 11.84AVTNPPVV271 pKa = 3.05
MM1 pKa = 7.83SSRR4 pKa = 11.84AWCFTLNNYY13 pKa = 9.94SEE15 pKa = 5.67DD16 pKa = 3.77DD17 pKa = 3.69LFEE20 pKa = 5.67ISGWDD25 pKa = 3.33CKK27 pKa = 10.6YY28 pKa = 10.38VVYY31 pKa = 10.14GVEE34 pKa = 4.13VAPEE38 pKa = 3.73TGTRR42 pKa = 11.84HH43 pKa = 4.76LQGYY47 pKa = 10.0CMFLGVQRR55 pKa = 11.84LSAVKK60 pKa = 10.24KK61 pKa = 10.43LLEE64 pKa = 4.09TAHH67 pKa = 6.32WEE69 pKa = 4.0IARR72 pKa = 11.84GSPLQNRR79 pKa = 11.84TYY81 pKa = 9.29CTKK84 pKa = 10.32EE85 pKa = 3.43GDD87 pKa = 3.78FVEE90 pKa = 5.21VGTLPLPRR98 pKa = 11.84GEE100 pKa = 4.29SEE102 pKa = 3.9KK103 pKa = 10.75EE104 pKa = 3.26RR105 pKa = 11.84WAEE108 pKa = 3.55AFEE111 pKa = 4.2AAKK114 pKa = 10.27HH115 pKa = 5.48GRR117 pKa = 11.84LDD119 pKa = 5.85DD120 pKa = 3.77IPEE123 pKa = 4.91DD124 pKa = 3.22IRR126 pKa = 11.84VRR128 pKa = 11.84YY129 pKa = 8.19YY130 pKa = 10.35RR131 pKa = 11.84TMKK134 pKa = 10.17EE135 pKa = 3.44IKK137 pKa = 8.78KK138 pKa = 9.97DD139 pKa = 3.59HH140 pKa = 5.62MVKK143 pKa = 10.6PEE145 pKa = 4.01DD146 pKa = 3.73QQDD149 pKa = 3.66VTGEE153 pKa = 3.9WFYY156 pKa = 11.85GEE158 pKa = 4.23PGVGKK163 pKa = 9.87SHH165 pKa = 7.95LARR168 pKa = 11.84EE169 pKa = 4.36QYY171 pKa = 10.04PGAYY175 pKa = 10.26LKK177 pKa = 9.28MQNKK181 pKa = 7.88WWDD184 pKa = 3.65GYY186 pKa = 10.21QDD188 pKa = 3.6EE189 pKa = 5.5KK190 pKa = 11.67YY191 pKa = 11.06VILDD195 pKa = 3.9DD196 pKa = 4.75FDD198 pKa = 5.07CKK200 pKa = 10.88EE201 pKa = 4.46LGHH204 pKa = 6.56HH205 pKa = 6.25LKK207 pKa = 9.94IWGDD211 pKa = 3.33KK212 pKa = 10.54YY213 pKa = 11.64SFLAEE218 pKa = 4.23TKK220 pKa = 10.47GGAICIRR227 pKa = 11.84PEE229 pKa = 3.84KK230 pKa = 10.78LIVTSNYY237 pKa = 9.74KK238 pKa = 10.2PEE240 pKa = 3.98QLFSDD245 pKa = 3.84EE246 pKa = 3.99MLIAVKK252 pKa = 10.35RR253 pKa = 11.84RR254 pKa = 11.84FKK256 pKa = 9.56VTHH259 pKa = 5.07VLSWRR264 pKa = 11.84AVTNPPVV271 pKa = 3.05
Molecular weight: 31.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI44|A0A0B4UI44_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-21 OX=1592088 PE=3 SV=1
MM1 pKa = 7.36VKK3 pKa = 10.03RR4 pKa = 11.84GYY6 pKa = 10.45GRR8 pKa = 11.84WRR10 pKa = 11.84AAAIGAGAAVVARR23 pKa = 11.84GARR26 pKa = 11.84YY27 pKa = 8.51AVKK30 pKa = 10.24RR31 pKa = 11.84VRR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.56MKK37 pKa = 10.16SRR39 pKa = 11.84PTVGRR44 pKa = 11.84KK45 pKa = 8.99RR46 pKa = 11.84KK47 pKa = 9.25MNMSKK52 pKa = 10.3PRR54 pKa = 11.84IAKK57 pKa = 9.61KK58 pKa = 10.39SRR60 pKa = 11.84GFGRR64 pKa = 11.84KK65 pKa = 8.64RR66 pKa = 11.84FGGLPNYY73 pKa = 10.11LNQGPGNEE81 pKa = 3.94LYY83 pKa = 9.69QWKK86 pKa = 8.17RR87 pKa = 11.84TARR90 pKa = 11.84IQRR93 pKa = 11.84PTLKK97 pKa = 10.39KK98 pKa = 9.9FLYY101 pKa = 6.78PTTISRR107 pKa = 11.84QDD109 pKa = 3.12RR110 pKa = 11.84FQGLTNFDD118 pKa = 3.92TNGGYY123 pKa = 10.71ARR125 pKa = 11.84LSYY128 pKa = 9.14HH129 pKa = 5.46TSIGEE134 pKa = 3.85NDD136 pKa = 3.96PRR138 pKa = 11.84WLPMAIFDD146 pKa = 4.13LTSFNNDD153 pKa = 2.74GAVPPYY159 pKa = 11.11SNVGYY164 pKa = 10.85KK165 pKa = 10.48LGWQNSGFGADD176 pKa = 3.69YY177 pKa = 10.7ARR179 pKa = 11.84SSIRR183 pKa = 11.84GQVPDD188 pKa = 4.65GDD190 pKa = 3.91ATTNTWTITSGTDD203 pKa = 2.76NGIYY207 pKa = 10.67NNISKK212 pKa = 10.56VYY214 pKa = 10.01LKK216 pKa = 10.38WIDD219 pKa = 3.09IRR221 pKa = 11.84LNLYY225 pKa = 8.66GARR228 pKa = 11.84KK229 pKa = 8.55RR230 pKa = 11.84TTKK233 pKa = 10.58FEE235 pKa = 4.01AQLVQFSNDD244 pKa = 2.93FGNLHH249 pKa = 6.54MGPLDD254 pKa = 3.85NPAGKK259 pKa = 10.22NNISYY264 pKa = 8.92MEE266 pKa = 4.38RR267 pKa = 11.84PLLFNNLQISTHH279 pKa = 6.16KK280 pKa = 8.94PEE282 pKa = 4.36KK283 pKa = 9.95MRR285 pKa = 11.84VLKK288 pKa = 10.05KK289 pKa = 9.79WKK291 pKa = 7.97WTVGPTTSIDD301 pKa = 3.71LNTTTGKK308 pKa = 9.56IQEE311 pKa = 4.16AKK313 pKa = 10.39IFLKK317 pKa = 10.13MNKK320 pKa = 9.64AINLDD325 pKa = 3.64YY326 pKa = 11.24SVGNNAVHH334 pKa = 6.61VDD336 pKa = 3.41VGDD339 pKa = 3.92GADD342 pKa = 3.63YY343 pKa = 11.29DD344 pKa = 3.86VDD346 pKa = 3.49TTTRR350 pKa = 11.84HH351 pKa = 6.63RR352 pKa = 11.84DD353 pKa = 3.44PKK355 pKa = 10.13PPSRR359 pKa = 11.84VYY361 pKa = 11.1LLLKK365 pKa = 10.12AFCPALTSLPSAALWDD381 pKa = 3.61KK382 pKa = 10.19TLAEE386 pKa = 4.56GSISNPFGGTALNNADD402 pKa = 3.92EE403 pKa = 4.56PSFDD407 pKa = 3.31FMIRR411 pKa = 11.84RR412 pKa = 11.84KK413 pKa = 10.04YY414 pKa = 10.69YY415 pKa = 9.92CDD417 pKa = 3.36KK418 pKa = 11.37
MM1 pKa = 7.36VKK3 pKa = 10.03RR4 pKa = 11.84GYY6 pKa = 10.45GRR8 pKa = 11.84WRR10 pKa = 11.84AAAIGAGAAVVARR23 pKa = 11.84GARR26 pKa = 11.84YY27 pKa = 8.51AVKK30 pKa = 10.24RR31 pKa = 11.84VRR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.56MKK37 pKa = 10.16SRR39 pKa = 11.84PTVGRR44 pKa = 11.84KK45 pKa = 8.99RR46 pKa = 11.84KK47 pKa = 9.25MNMSKK52 pKa = 10.3PRR54 pKa = 11.84IAKK57 pKa = 9.61KK58 pKa = 10.39SRR60 pKa = 11.84GFGRR64 pKa = 11.84KK65 pKa = 8.64RR66 pKa = 11.84FGGLPNYY73 pKa = 10.11LNQGPGNEE81 pKa = 3.94LYY83 pKa = 9.69QWKK86 pKa = 8.17RR87 pKa = 11.84TARR90 pKa = 11.84IQRR93 pKa = 11.84PTLKK97 pKa = 10.39KK98 pKa = 9.9FLYY101 pKa = 6.78PTTISRR107 pKa = 11.84QDD109 pKa = 3.12RR110 pKa = 11.84FQGLTNFDD118 pKa = 3.92TNGGYY123 pKa = 10.71ARR125 pKa = 11.84LSYY128 pKa = 9.14HH129 pKa = 5.46TSIGEE134 pKa = 3.85NDD136 pKa = 3.96PRR138 pKa = 11.84WLPMAIFDD146 pKa = 4.13LTSFNNDD153 pKa = 2.74GAVPPYY159 pKa = 11.11SNVGYY164 pKa = 10.85KK165 pKa = 10.48LGWQNSGFGADD176 pKa = 3.69YY177 pKa = 10.7ARR179 pKa = 11.84SSIRR183 pKa = 11.84GQVPDD188 pKa = 4.65GDD190 pKa = 3.91ATTNTWTITSGTDD203 pKa = 2.76NGIYY207 pKa = 10.67NNISKK212 pKa = 10.56VYY214 pKa = 10.01LKK216 pKa = 10.38WIDD219 pKa = 3.09IRR221 pKa = 11.84LNLYY225 pKa = 8.66GARR228 pKa = 11.84KK229 pKa = 8.55RR230 pKa = 11.84TTKK233 pKa = 10.58FEE235 pKa = 4.01AQLVQFSNDD244 pKa = 2.93FGNLHH249 pKa = 6.54MGPLDD254 pKa = 3.85NPAGKK259 pKa = 10.22NNISYY264 pKa = 8.92MEE266 pKa = 4.38RR267 pKa = 11.84PLLFNNLQISTHH279 pKa = 6.16KK280 pKa = 8.94PEE282 pKa = 4.36KK283 pKa = 9.95MRR285 pKa = 11.84VLKK288 pKa = 10.05KK289 pKa = 9.79WKK291 pKa = 7.97WTVGPTTSIDD301 pKa = 3.71LNTTTGKK308 pKa = 9.56IQEE311 pKa = 4.16AKK313 pKa = 10.39IFLKK317 pKa = 10.13MNKK320 pKa = 9.64AINLDD325 pKa = 3.64YY326 pKa = 11.24SVGNNAVHH334 pKa = 6.61VDD336 pKa = 3.41VGDD339 pKa = 3.92GADD342 pKa = 3.63YY343 pKa = 11.29DD344 pKa = 3.86VDD346 pKa = 3.49TTTRR350 pKa = 11.84HH351 pKa = 6.63RR352 pKa = 11.84DD353 pKa = 3.44PKK355 pKa = 10.13PPSRR359 pKa = 11.84VYY361 pKa = 11.1LLLKK365 pKa = 10.12AFCPALTSLPSAALWDD381 pKa = 3.61KK382 pKa = 10.19TLAEE386 pKa = 4.56GSISNPFGGTALNNADD402 pKa = 3.92EE403 pKa = 4.56PSFDD407 pKa = 3.31FMIRR411 pKa = 11.84RR412 pKa = 11.84KK413 pKa = 10.04YY414 pKa = 10.69YY415 pKa = 9.92CDD417 pKa = 3.36KK418 pKa = 11.37
Molecular weight: 47.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
689 |
271 |
418 |
344.5 |
39.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.676 ± 0.674 | 1.161 ± 0.622 |
5.951 ± 0.19 | 4.79 ± 2.619 |
3.919 ± 0.135 | 8.273 ± 0.527 |
1.887 ± 0.629 | 4.499 ± 0.26 |
7.837 ± 0.166 | 7.547 ± 0.119 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.322 ± 0.064 | 5.37 ± 1.864 |
5.08 ± 0.385 | 2.903 ± 0.247 |
7.257 ± 0.799 | 5.225 ± 0.471 |
6.241 ± 0.853 | 5.515 ± 0.883 |
2.612 ± 0.418 | 4.935 ± 0.137 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
