
Biratnagar virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
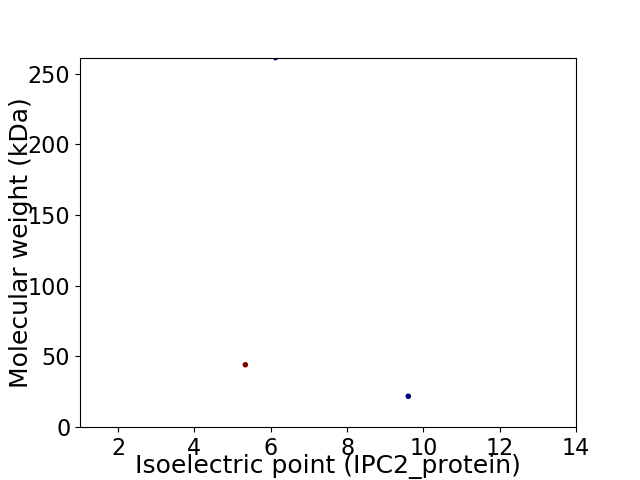
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7EIZ3|A0A1U7EIZ3_9VIRU Non-structural protein 2 OS=Biratnagar virus OX=1955197 PE=4 SV=1
MM1 pKa = 7.18FLIVFLFSTAFAFDD15 pKa = 4.7RR16 pKa = 11.84YY17 pKa = 9.46PLPEE21 pKa = 3.99LVEE24 pKa = 4.18FHH26 pKa = 7.18SEE28 pKa = 3.8LDD30 pKa = 3.42RR31 pKa = 11.84LGMLDD36 pKa = 3.41IEE38 pKa = 5.51LIGKK42 pKa = 9.41LYY44 pKa = 10.61SLQPFSDD51 pKa = 3.85FKK53 pKa = 11.43VYY55 pKa = 9.7EE56 pKa = 4.01ANAWNKK62 pKa = 10.39LIGYY66 pKa = 7.34HH67 pKa = 6.19CPSGQTVYY75 pKa = 10.59PLEE78 pKa = 4.33HH79 pKa = 6.78SFLKK83 pKa = 10.61DD84 pKa = 3.79LYY86 pKa = 10.26TCIPKK91 pKa = 10.29PKK93 pKa = 9.95VLNSQYY99 pKa = 10.84LVYY102 pKa = 10.42RR103 pKa = 11.84IKK105 pKa = 10.65PSSHH109 pKa = 7.26DD110 pKa = 3.69NVTVQCKK117 pKa = 8.39NRR119 pKa = 11.84YY120 pKa = 8.24YY121 pKa = 9.77NVSVAFCQIEE131 pKa = 4.58SYY133 pKa = 10.97RR134 pKa = 11.84FLIDD138 pKa = 3.44NKK140 pKa = 10.04GDD142 pKa = 3.07TWFTSDD148 pKa = 3.24YY149 pKa = 9.85CLSNRR154 pKa = 11.84YY155 pKa = 7.22MTDD158 pKa = 2.96NDD160 pKa = 3.69GKK162 pKa = 10.58EE163 pKa = 4.2LFTVSHH169 pKa = 6.95TSTDD173 pKa = 3.44YY174 pKa = 11.29SVTFEE179 pKa = 4.63FPPPEE184 pKa = 4.49CPFYY188 pKa = 10.48PYY190 pKa = 10.73DD191 pKa = 3.46VYY193 pKa = 11.43YY194 pKa = 10.52EE195 pKa = 4.14KK196 pKa = 10.81SLPATVCIDD205 pKa = 3.45SSPEE209 pKa = 3.73FKK211 pKa = 10.81SCVTGAKK218 pKa = 10.17CEE220 pKa = 3.94VHH222 pKa = 6.6PPYY225 pKa = 10.69EE226 pKa = 4.11LLPVSTHH233 pKa = 6.15SEE235 pKa = 3.72LHH237 pKa = 5.93FKK239 pKa = 11.09VNGTCSVAFMTFTEE253 pKa = 4.55PTTITYY259 pKa = 9.59HH260 pKa = 5.99IKK262 pKa = 10.47SALSTSFIDD271 pKa = 4.33AVLHH275 pKa = 6.47ALLSLLRR282 pKa = 11.84PLIDD286 pKa = 4.22ALLEE290 pKa = 4.03LVTYY294 pKa = 10.43IIDD297 pKa = 3.65TLVTIFTSSEE307 pKa = 3.62FVAIYY312 pKa = 9.91QKK314 pKa = 10.18IYY316 pKa = 10.93HH317 pKa = 7.24LILSVLQNVFDD328 pKa = 4.46YY329 pKa = 10.67IIDD332 pKa = 3.76VAVPKK337 pKa = 10.73IMDD340 pKa = 4.58LFYY343 pKa = 11.16EE344 pKa = 4.51STLKK348 pKa = 10.13TKK350 pKa = 10.32FIILTFIYY358 pKa = 10.6VYY360 pKa = 10.46FKK362 pKa = 9.32YY363 pKa = 9.51TKK365 pKa = 10.22FIYY368 pKa = 10.24ALSVCVVVCLFFSKK382 pKa = 10.89
MM1 pKa = 7.18FLIVFLFSTAFAFDD15 pKa = 4.7RR16 pKa = 11.84YY17 pKa = 9.46PLPEE21 pKa = 3.99LVEE24 pKa = 4.18FHH26 pKa = 7.18SEE28 pKa = 3.8LDD30 pKa = 3.42RR31 pKa = 11.84LGMLDD36 pKa = 3.41IEE38 pKa = 5.51LIGKK42 pKa = 9.41LYY44 pKa = 10.61SLQPFSDD51 pKa = 3.85FKK53 pKa = 11.43VYY55 pKa = 9.7EE56 pKa = 4.01ANAWNKK62 pKa = 10.39LIGYY66 pKa = 7.34HH67 pKa = 6.19CPSGQTVYY75 pKa = 10.59PLEE78 pKa = 4.33HH79 pKa = 6.78SFLKK83 pKa = 10.61DD84 pKa = 3.79LYY86 pKa = 10.26TCIPKK91 pKa = 10.29PKK93 pKa = 9.95VLNSQYY99 pKa = 10.84LVYY102 pKa = 10.42RR103 pKa = 11.84IKK105 pKa = 10.65PSSHH109 pKa = 7.26DD110 pKa = 3.69NVTVQCKK117 pKa = 8.39NRR119 pKa = 11.84YY120 pKa = 8.24YY121 pKa = 9.77NVSVAFCQIEE131 pKa = 4.58SYY133 pKa = 10.97RR134 pKa = 11.84FLIDD138 pKa = 3.44NKK140 pKa = 10.04GDD142 pKa = 3.07TWFTSDD148 pKa = 3.24YY149 pKa = 9.85CLSNRR154 pKa = 11.84YY155 pKa = 7.22MTDD158 pKa = 2.96NDD160 pKa = 3.69GKK162 pKa = 10.58EE163 pKa = 4.2LFTVSHH169 pKa = 6.95TSTDD173 pKa = 3.44YY174 pKa = 11.29SVTFEE179 pKa = 4.63FPPPEE184 pKa = 4.49CPFYY188 pKa = 10.48PYY190 pKa = 10.73DD191 pKa = 3.46VYY193 pKa = 11.43YY194 pKa = 10.52EE195 pKa = 4.14KK196 pKa = 10.81SLPATVCIDD205 pKa = 3.45SSPEE209 pKa = 3.73FKK211 pKa = 10.81SCVTGAKK218 pKa = 10.17CEE220 pKa = 3.94VHH222 pKa = 6.6PPYY225 pKa = 10.69EE226 pKa = 4.11LLPVSTHH233 pKa = 6.15SEE235 pKa = 3.72LHH237 pKa = 5.93FKK239 pKa = 11.09VNGTCSVAFMTFTEE253 pKa = 4.55PTTITYY259 pKa = 9.59HH260 pKa = 5.99IKK262 pKa = 10.47SALSTSFIDD271 pKa = 4.33AVLHH275 pKa = 6.47ALLSLLRR282 pKa = 11.84PLIDD286 pKa = 4.22ALLEE290 pKa = 4.03LVTYY294 pKa = 10.43IIDD297 pKa = 3.65TLVTIFTSSEE307 pKa = 3.62FVAIYY312 pKa = 9.91QKK314 pKa = 10.18IYY316 pKa = 10.93HH317 pKa = 7.24LILSVLQNVFDD328 pKa = 4.46YY329 pKa = 10.67IIDD332 pKa = 3.76VAVPKK337 pKa = 10.73IMDD340 pKa = 4.58LFYY343 pKa = 11.16EE344 pKa = 4.51STLKK348 pKa = 10.13TKK350 pKa = 10.32FIILTFIYY358 pKa = 10.6VYY360 pKa = 10.46FKK362 pKa = 9.32YY363 pKa = 9.51TKK365 pKa = 10.22FIYY368 pKa = 10.24ALSVCVVVCLFFSKK382 pKa = 10.89
Molecular weight: 44.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7EIU8|A0A1U7EIU8_9VIRU Uncharacterized protein OS=Biratnagar virus OX=1955197 PE=4 SV=1
MM1 pKa = 6.82VLKK4 pKa = 9.91RR5 pKa = 11.84TGTVVRR11 pKa = 11.84PTTTSRR17 pKa = 11.84RR18 pKa = 11.84AVVAAPVKK26 pKa = 9.97RR27 pKa = 11.84AVNRR31 pKa = 11.84VKK33 pKa = 11.03NFGSNLDD40 pKa = 3.5FSKK43 pKa = 10.61YY44 pKa = 8.17FQYY47 pKa = 10.13ATSVLSNTTFLLIFAFSAFLCYY69 pKa = 10.64NYY71 pKa = 7.56TTKK74 pKa = 10.91GEE76 pKa = 3.99KK77 pKa = 9.37SHH79 pKa = 6.24IVKK82 pKa = 10.26FVGNFVTQFPSFKK95 pKa = 10.55DD96 pKa = 3.63SEE98 pKa = 4.3CSILGFVQFFVPFIPAVVSVRR119 pKa = 11.84PSNRR123 pKa = 11.84VATLVGVALYY133 pKa = 10.39YY134 pKa = 10.71VFVPEE139 pKa = 4.11RR140 pKa = 11.84TVYY143 pKa = 10.13EE144 pKa = 4.18YY145 pKa = 10.75LVHH148 pKa = 6.97GIIMYY153 pKa = 10.24LILRR157 pKa = 11.84TEE159 pKa = 3.8VRR161 pKa = 11.84AYY163 pKa = 10.78RR164 pKa = 11.84LIGFAMLFLSYY175 pKa = 10.55IMQFALPLPSGDD187 pKa = 3.28GHH189 pKa = 6.58NCTSSS194 pKa = 2.75
MM1 pKa = 6.82VLKK4 pKa = 9.91RR5 pKa = 11.84TGTVVRR11 pKa = 11.84PTTTSRR17 pKa = 11.84RR18 pKa = 11.84AVVAAPVKK26 pKa = 9.97RR27 pKa = 11.84AVNRR31 pKa = 11.84VKK33 pKa = 11.03NFGSNLDD40 pKa = 3.5FSKK43 pKa = 10.61YY44 pKa = 8.17FQYY47 pKa = 10.13ATSVLSNTTFLLIFAFSAFLCYY69 pKa = 10.64NYY71 pKa = 7.56TTKK74 pKa = 10.91GEE76 pKa = 3.99KK77 pKa = 9.37SHH79 pKa = 6.24IVKK82 pKa = 10.26FVGNFVTQFPSFKK95 pKa = 10.55DD96 pKa = 3.63SEE98 pKa = 4.3CSILGFVQFFVPFIPAVVSVRR119 pKa = 11.84PSNRR123 pKa = 11.84VATLVGVALYY133 pKa = 10.39YY134 pKa = 10.71VFVPEE139 pKa = 4.11RR140 pKa = 11.84TVYY143 pKa = 10.13EE144 pKa = 4.18YY145 pKa = 10.75LVHH148 pKa = 6.97GIIMYY153 pKa = 10.24LILRR157 pKa = 11.84TEE159 pKa = 3.8VRR161 pKa = 11.84AYY163 pKa = 10.78RR164 pKa = 11.84LIGFAMLFLSYY175 pKa = 10.55IMQFALPLPSGDD187 pKa = 3.28GHH189 pKa = 6.58NCTSSS194 pKa = 2.75
Molecular weight: 21.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2841 |
194 |
2265 |
947.0 |
109.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.857 ± 0.508 | 2.358 ± 0.284 |
6.617 ± 1.59 | 4.963 ± 0.657 |
6.16 ± 1.335 | 2.957 ± 0.586 |
2.851 ± 0.357 | 7.005 ± 0.674 |
6.688 ± 0.847 | 9.187 ± 0.561 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.31 | 4.717 ± 0.662 |
4.4 ± 0.38 | 2.64 ± 0.344 |
4.435 ± 0.836 | 7.673 ± 0.39 |
6.512 ± 0.553 | 7.779 ± 1.477 |
0.422 ± 0.111 | 5.561 ± 0.667 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
