
Maize associated rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
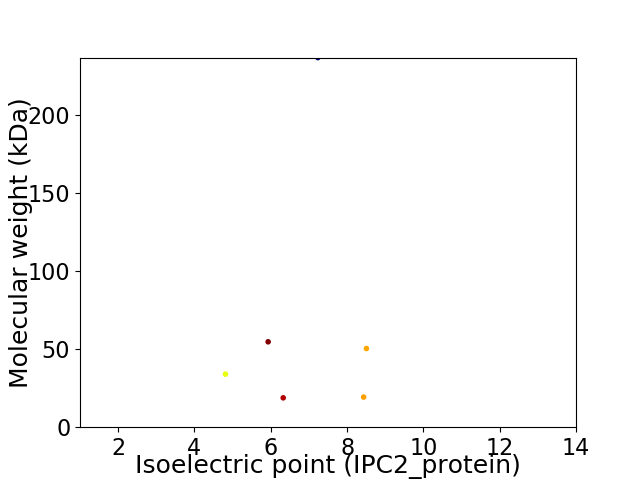
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9Y2V8|A0A1X9Y2V8_9RHAB Putative P protein OS=Maize associated rhabdovirus OX=2003308 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 5.23APKK5 pKa = 10.69ASDD8 pKa = 3.32IFSQVPDD15 pKa = 4.2DD16 pKa = 4.21VEE18 pKa = 4.94PDD20 pKa = 3.57YY21 pKa = 11.36EE22 pKa = 4.23IDD24 pKa = 3.45EE25 pKa = 4.87DD26 pKa = 4.97EE27 pKa = 4.86IFAKK31 pKa = 10.4PGPTQAEE38 pKa = 4.29PNNQEE43 pKa = 3.92DD44 pKa = 4.01ALDD47 pKa = 3.86EE48 pKa = 4.38EE49 pKa = 5.23EE50 pKa = 5.01EE51 pKa = 4.41EE52 pKa = 4.36QHH54 pKa = 6.61SDD56 pKa = 3.49PPPASGNPPDD66 pKa = 4.37YY67 pKa = 10.62RR68 pKa = 11.84FKK70 pKa = 10.57TISAMTDD77 pKa = 3.73FVRR80 pKa = 11.84KK81 pKa = 7.04TANDD85 pKa = 3.11KK86 pKa = 11.37GITVKK91 pKa = 10.77KK92 pKa = 9.21EE93 pKa = 3.34WMNILTRR100 pKa = 11.84EE101 pKa = 4.03WHH103 pKa = 6.43KK104 pKa = 11.53KK105 pKa = 8.67NADD108 pKa = 3.33VYY110 pKa = 10.96ASHH113 pKa = 7.56IDD115 pKa = 3.72FFFLGITGEE124 pKa = 4.31RR125 pKa = 11.84NLSVEE130 pKa = 4.64KK131 pKa = 10.66DD132 pKa = 3.61FKK134 pKa = 11.39DD135 pKa = 2.87IAKK138 pKa = 10.4RR139 pKa = 11.84MNDD142 pKa = 3.14DD143 pKa = 3.43VNRR146 pKa = 11.84VSGVGKK152 pKa = 8.55QLKK155 pKa = 10.13DD156 pKa = 3.29ISDD159 pKa = 3.87KK160 pKa = 10.5MSKK163 pKa = 10.73DD164 pKa = 3.35LDD166 pKa = 3.73AKK168 pKa = 10.46MKK170 pKa = 10.85LLDD173 pKa = 4.07AKK175 pKa = 10.71LEE177 pKa = 4.26KK178 pKa = 10.05FNSMMEE184 pKa = 4.17KK185 pKa = 10.37NEE187 pKa = 4.02SFLEE191 pKa = 4.32SASKK195 pKa = 10.19QSSVASWAAEE205 pKa = 3.79ARR207 pKa = 11.84DD208 pKa = 4.14EE209 pKa = 4.31PPKK212 pKa = 10.15TKK214 pKa = 9.22KK215 pKa = 8.87TSPRR219 pKa = 11.84MKK221 pKa = 9.84ILRR224 pKa = 11.84FLSGIGFSQVEE235 pKa = 4.25AQNVIKK241 pKa = 10.82VGAHH245 pKa = 6.11HH246 pKa = 7.49IITEE250 pKa = 4.03EE251 pKa = 3.7MMEE254 pKa = 4.15NVFTGLADD262 pKa = 4.35DD263 pKa = 4.31EE264 pKa = 4.85LKK266 pKa = 11.0EE267 pKa = 4.29EE268 pKa = 4.36YY269 pKa = 10.4EE270 pKa = 4.7QMLCLEE276 pKa = 4.29GQKK279 pKa = 10.48LVAEE283 pKa = 4.34ARR285 pKa = 11.84EE286 pKa = 4.17KK287 pKa = 10.96KK288 pKa = 10.1NKK290 pKa = 10.14KK291 pKa = 9.36KK292 pKa = 10.63DD293 pKa = 3.2IYY295 pKa = 11.09DD296 pKa = 4.02DD297 pKa = 3.92YY298 pKa = 12.15
MM1 pKa = 8.09DD2 pKa = 5.23APKK5 pKa = 10.69ASDD8 pKa = 3.32IFSQVPDD15 pKa = 4.2DD16 pKa = 4.21VEE18 pKa = 4.94PDD20 pKa = 3.57YY21 pKa = 11.36EE22 pKa = 4.23IDD24 pKa = 3.45EE25 pKa = 4.87DD26 pKa = 4.97EE27 pKa = 4.86IFAKK31 pKa = 10.4PGPTQAEE38 pKa = 4.29PNNQEE43 pKa = 3.92DD44 pKa = 4.01ALDD47 pKa = 3.86EE48 pKa = 4.38EE49 pKa = 5.23EE50 pKa = 5.01EE51 pKa = 4.41EE52 pKa = 4.36QHH54 pKa = 6.61SDD56 pKa = 3.49PPPASGNPPDD66 pKa = 4.37YY67 pKa = 10.62RR68 pKa = 11.84FKK70 pKa = 10.57TISAMTDD77 pKa = 3.73FVRR80 pKa = 11.84KK81 pKa = 7.04TANDD85 pKa = 3.11KK86 pKa = 11.37GITVKK91 pKa = 10.77KK92 pKa = 9.21EE93 pKa = 3.34WMNILTRR100 pKa = 11.84EE101 pKa = 4.03WHH103 pKa = 6.43KK104 pKa = 11.53KK105 pKa = 8.67NADD108 pKa = 3.33VYY110 pKa = 10.96ASHH113 pKa = 7.56IDD115 pKa = 3.72FFFLGITGEE124 pKa = 4.31RR125 pKa = 11.84NLSVEE130 pKa = 4.64KK131 pKa = 10.66DD132 pKa = 3.61FKK134 pKa = 11.39DD135 pKa = 2.87IAKK138 pKa = 10.4RR139 pKa = 11.84MNDD142 pKa = 3.14DD143 pKa = 3.43VNRR146 pKa = 11.84VSGVGKK152 pKa = 8.55QLKK155 pKa = 10.13DD156 pKa = 3.29ISDD159 pKa = 3.87KK160 pKa = 10.5MSKK163 pKa = 10.73DD164 pKa = 3.35LDD166 pKa = 3.73AKK168 pKa = 10.46MKK170 pKa = 10.85LLDD173 pKa = 4.07AKK175 pKa = 10.71LEE177 pKa = 4.26KK178 pKa = 10.05FNSMMEE184 pKa = 4.17KK185 pKa = 10.37NEE187 pKa = 4.02SFLEE191 pKa = 4.32SASKK195 pKa = 10.19QSSVASWAAEE205 pKa = 3.79ARR207 pKa = 11.84DD208 pKa = 4.14EE209 pKa = 4.31PPKK212 pKa = 10.15TKK214 pKa = 9.22KK215 pKa = 8.87TSPRR219 pKa = 11.84MKK221 pKa = 9.84ILRR224 pKa = 11.84FLSGIGFSQVEE235 pKa = 4.25AQNVIKK241 pKa = 10.82VGAHH245 pKa = 6.11HH246 pKa = 7.49IITEE250 pKa = 4.03EE251 pKa = 3.7MMEE254 pKa = 4.15NVFTGLADD262 pKa = 4.35DD263 pKa = 4.31EE264 pKa = 4.85LKK266 pKa = 11.0EE267 pKa = 4.29EE268 pKa = 4.36YY269 pKa = 10.4EE270 pKa = 4.7QMLCLEE276 pKa = 4.29GQKK279 pKa = 10.48LVAEE283 pKa = 4.34ARR285 pKa = 11.84EE286 pKa = 4.17KK287 pKa = 10.96KK288 pKa = 10.1NKK290 pKa = 10.14KK291 pKa = 9.36KK292 pKa = 10.63DD293 pKa = 3.2IYY295 pKa = 11.09DD296 pKa = 4.02DD297 pKa = 3.92YY298 pKa = 12.15
Molecular weight: 33.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9Y2V3|A0A1X9Y2V3_9RHAB Putative G protein OS=Maize associated rhabdovirus OX=2003308 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84PRR4 pKa = 11.84SLGFLHH10 pKa = 7.18LRR12 pKa = 11.84ILRR15 pKa = 11.84VKK17 pKa = 8.65MASGSEE23 pKa = 3.76QHH25 pKa = 6.59IDD27 pKa = 4.24KK28 pKa = 10.91DD29 pKa = 3.66LTDD32 pKa = 3.99AFSNVPEE39 pKa = 4.06NLAVNTVPDD48 pKa = 3.74EE49 pKa = 4.33TFSDD53 pKa = 4.05DD54 pKa = 3.2AFKK57 pKa = 10.97SHH59 pKa = 6.35SVYY62 pKa = 10.51EE63 pKa = 4.26INEE66 pKa = 3.97LTIKK70 pKa = 10.47QVVHH74 pKa = 6.17HH75 pKa = 7.08AYY77 pKa = 10.02KK78 pKa = 10.83LFRR81 pKa = 11.84NMGQKK86 pKa = 8.24TCHH89 pKa = 6.5AIIPYY94 pKa = 10.05SILAMALRR102 pKa = 11.84LPTPKK107 pKa = 10.17KK108 pKa = 10.38KK109 pKa = 9.47STTSFLTSHH118 pKa = 6.58KK119 pKa = 9.86PSGGTALEE127 pKa = 3.71QGIASKK133 pKa = 10.64LYY135 pKa = 9.72PSLDD139 pKa = 3.37LSTEE143 pKa = 3.95EE144 pKa = 4.06AAKK147 pKa = 10.32PDD149 pKa = 3.77AEE151 pKa = 4.52KK152 pKa = 10.93KK153 pKa = 10.39RR154 pKa = 11.84LTATKK159 pKa = 10.41VFGSEE164 pKa = 3.44EE165 pKa = 4.02DD166 pKa = 3.75KK167 pKa = 11.01EE168 pKa = 4.61FKK170 pKa = 10.38AVRR173 pKa = 11.84EE174 pKa = 4.09LEE176 pKa = 4.13EE177 pKa = 4.54EE178 pKa = 3.97IVFQFGCFLSAFLLKK193 pKa = 10.35LLCKK197 pKa = 10.37NPEE200 pKa = 4.15NIITGWDD207 pKa = 3.36AMRR210 pKa = 11.84GRR212 pKa = 11.84YY213 pKa = 7.63STFFSEE219 pKa = 4.64SAPSEE224 pKa = 4.13VKK226 pKa = 10.38RR227 pKa = 11.84PSRR230 pKa = 11.84EE231 pKa = 3.43WLTEE235 pKa = 3.74LKK237 pKa = 10.78NLFSSDD243 pKa = 3.23PMISRR248 pKa = 11.84TWVRR252 pKa = 11.84TFSEE256 pKa = 4.9AEE258 pKa = 3.77DD259 pKa = 3.7HH260 pKa = 7.0LPAGEE265 pKa = 4.12QQVGMLRR272 pKa = 11.84YY273 pKa = 8.84LALMPFSYY281 pKa = 10.03TGLHH285 pKa = 6.43GYY287 pKa = 10.42KK288 pKa = 10.4LFLDD292 pKa = 4.0VSRR295 pKa = 11.84ASNLSNKK302 pKa = 8.64WLLTEE307 pKa = 4.11MTSPMTSKK315 pKa = 10.76ALKK318 pKa = 10.15LIAHH322 pKa = 6.91ILINFEE328 pKa = 4.02SRR330 pKa = 11.84IGEE333 pKa = 4.23KK334 pKa = 10.19KK335 pKa = 10.33KK336 pKa = 10.93SAFKK340 pKa = 9.39YY341 pKa = 9.87ARR343 pKa = 11.84MMSTSYY349 pKa = 10.77FQDD352 pKa = 3.44LQTKK356 pKa = 8.21NCAGLVYY363 pKa = 10.8LEE365 pKa = 4.02VKK367 pKa = 9.89ILKK370 pKa = 10.09KK371 pKa = 10.69FEE373 pKa = 4.34AFGTNSNPEE382 pKa = 4.1NIIGIEE388 pKa = 4.03KK389 pKa = 10.37VPAEE393 pKa = 4.32HH394 pKa = 7.58KK395 pKa = 11.21AMLSKK400 pKa = 10.73AADD403 pKa = 4.54FIVSAAPQRR412 pKa = 11.84NLGRR416 pKa = 11.84YY417 pKa = 8.31SEE419 pKa = 4.41SMRR422 pKa = 11.84KK423 pKa = 9.9AMIPEE428 pKa = 4.66HH429 pKa = 6.79KK430 pKa = 9.62PKK432 pKa = 10.17QHH434 pKa = 4.92QQVAKK439 pKa = 10.5KK440 pKa = 10.25EE441 pKa = 4.43DD442 pKa = 3.3IFGG445 pKa = 3.8
MM1 pKa = 7.97RR2 pKa = 11.84PRR4 pKa = 11.84SLGFLHH10 pKa = 7.18LRR12 pKa = 11.84ILRR15 pKa = 11.84VKK17 pKa = 8.65MASGSEE23 pKa = 3.76QHH25 pKa = 6.59IDD27 pKa = 4.24KK28 pKa = 10.91DD29 pKa = 3.66LTDD32 pKa = 3.99AFSNVPEE39 pKa = 4.06NLAVNTVPDD48 pKa = 3.74EE49 pKa = 4.33TFSDD53 pKa = 4.05DD54 pKa = 3.2AFKK57 pKa = 10.97SHH59 pKa = 6.35SVYY62 pKa = 10.51EE63 pKa = 4.26INEE66 pKa = 3.97LTIKK70 pKa = 10.47QVVHH74 pKa = 6.17HH75 pKa = 7.08AYY77 pKa = 10.02KK78 pKa = 10.83LFRR81 pKa = 11.84NMGQKK86 pKa = 8.24TCHH89 pKa = 6.5AIIPYY94 pKa = 10.05SILAMALRR102 pKa = 11.84LPTPKK107 pKa = 10.17KK108 pKa = 10.38KK109 pKa = 9.47STTSFLTSHH118 pKa = 6.58KK119 pKa = 9.86PSGGTALEE127 pKa = 3.71QGIASKK133 pKa = 10.64LYY135 pKa = 9.72PSLDD139 pKa = 3.37LSTEE143 pKa = 3.95EE144 pKa = 4.06AAKK147 pKa = 10.32PDD149 pKa = 3.77AEE151 pKa = 4.52KK152 pKa = 10.93KK153 pKa = 10.39RR154 pKa = 11.84LTATKK159 pKa = 10.41VFGSEE164 pKa = 3.44EE165 pKa = 4.02DD166 pKa = 3.75KK167 pKa = 11.01EE168 pKa = 4.61FKK170 pKa = 10.38AVRR173 pKa = 11.84EE174 pKa = 4.09LEE176 pKa = 4.13EE177 pKa = 4.54EE178 pKa = 3.97IVFQFGCFLSAFLLKK193 pKa = 10.35LLCKK197 pKa = 10.37NPEE200 pKa = 4.15NIITGWDD207 pKa = 3.36AMRR210 pKa = 11.84GRR212 pKa = 11.84YY213 pKa = 7.63STFFSEE219 pKa = 4.64SAPSEE224 pKa = 4.13VKK226 pKa = 10.38RR227 pKa = 11.84PSRR230 pKa = 11.84EE231 pKa = 3.43WLTEE235 pKa = 3.74LKK237 pKa = 10.78NLFSSDD243 pKa = 3.23PMISRR248 pKa = 11.84TWVRR252 pKa = 11.84TFSEE256 pKa = 4.9AEE258 pKa = 3.77DD259 pKa = 3.7HH260 pKa = 7.0LPAGEE265 pKa = 4.12QQVGMLRR272 pKa = 11.84YY273 pKa = 8.84LALMPFSYY281 pKa = 10.03TGLHH285 pKa = 6.43GYY287 pKa = 10.42KK288 pKa = 10.4LFLDD292 pKa = 4.0VSRR295 pKa = 11.84ASNLSNKK302 pKa = 8.64WLLTEE307 pKa = 4.11MTSPMTSKK315 pKa = 10.76ALKK318 pKa = 10.15LIAHH322 pKa = 6.91ILINFEE328 pKa = 4.02SRR330 pKa = 11.84IGEE333 pKa = 4.23KK334 pKa = 10.19KK335 pKa = 10.33KK336 pKa = 10.93SAFKK340 pKa = 9.39YY341 pKa = 9.87ARR343 pKa = 11.84MMSTSYY349 pKa = 10.77FQDD352 pKa = 3.44LQTKK356 pKa = 8.21NCAGLVYY363 pKa = 10.8LEE365 pKa = 4.02VKK367 pKa = 9.89ILKK370 pKa = 10.09KK371 pKa = 10.69FEE373 pKa = 4.34AFGTNSNPEE382 pKa = 4.1NIIGIEE388 pKa = 4.03KK389 pKa = 10.37VPAEE393 pKa = 4.32HH394 pKa = 7.58KK395 pKa = 11.21AMLSKK400 pKa = 10.73AADD403 pKa = 4.54FIVSAAPQRR412 pKa = 11.84NLGRR416 pKa = 11.84YY417 pKa = 8.31SEE419 pKa = 4.41SMRR422 pKa = 11.84KK423 pKa = 9.9AMIPEE428 pKa = 4.66HH429 pKa = 6.79KK430 pKa = 9.62PKK432 pKa = 10.17QHH434 pKa = 4.92QQVAKK439 pKa = 10.5KK440 pKa = 10.25EE441 pKa = 4.43DD442 pKa = 3.3IFGG445 pKa = 3.8
Molecular weight: 50.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3631 |
167 |
2066 |
605.2 |
68.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.426 ± 0.682 | 1.708 ± 0.703 |
5.095 ± 0.805 | 6.995 ± 0.875 |
4.241 ± 0.427 | 5.288 ± 0.417 |
2.368 ± 0.128 | 8.014 ± 0.786 |
6.968 ± 1.083 | 9.226 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.167 ± 0.306 | 4.351 ± 0.132 |
4.875 ± 0.054 | 2.809 ± 0.337 |
5.095 ± 0.618 | 8.207 ± 0.566 |
5.839 ± 0.409 | 5.095 ± 0.665 |
1.652 ± 0.374 | 3.58 ± 0.429 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
