
Anaerococcus lactolyticus ATCC 51172
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellales; Peptoniphilaceae; Anaerococcus; Anaerococcus lactolyticus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
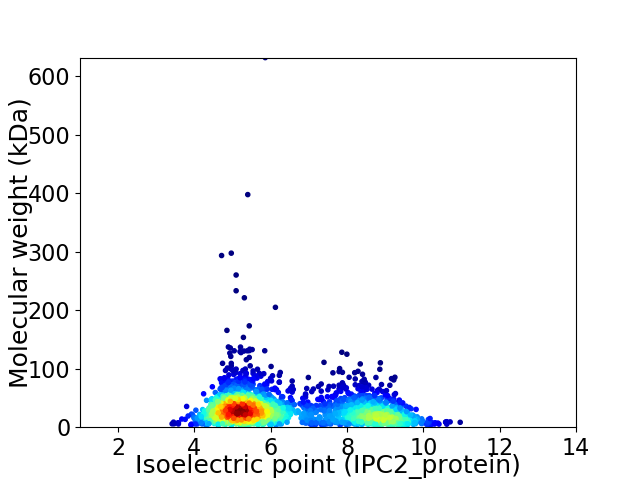
Virtual 2D-PAGE plot for 2252 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C2BDB1|C2BDB1_9FIRM Radical SAM protein TIGR01212 family OS=Anaerococcus lactolyticus ATCC 51172 OX=525254 GN=HMPREF0072_0331 PE=4 SV=1
MM1 pKa = 7.56SYY3 pKa = 11.03YY4 pKa = 10.25EE5 pKa = 3.7VRR7 pKa = 11.84ANVNQLISEE16 pKa = 4.18NYY18 pKa = 7.77EE19 pKa = 4.08DD20 pKa = 4.18YY21 pKa = 11.09VKK23 pKa = 11.07SLISIEE29 pKa = 4.21KK30 pKa = 10.03NINNEE35 pKa = 3.77DD36 pKa = 4.15CLNSIYY42 pKa = 11.06NEE44 pKa = 4.52FINDD48 pKa = 3.18NNAASIFDD56 pKa = 4.21ADD58 pKa = 3.04IDD60 pKa = 4.42GYY62 pKa = 10.22IEE64 pKa = 4.13KK65 pKa = 10.62LGIDD69 pKa = 3.94LEE71 pKa = 4.46DD72 pKa = 3.35EE73 pKa = 4.27RR74 pKa = 11.84VIHH77 pKa = 6.32GEE79 pKa = 3.71PVSYY83 pKa = 10.81EE84 pKa = 3.91EE85 pKa = 4.01EE86 pKa = 4.0QEE88 pKa = 4.31RR89 pKa = 11.84EE90 pKa = 3.51VDD92 pKa = 3.25RR93 pKa = 11.84EE94 pKa = 4.03KK95 pKa = 11.11FNKK98 pKa = 9.52NDD100 pKa = 3.63YY101 pKa = 10.86EE102 pKa = 4.15NLKK105 pKa = 10.14PDD107 pKa = 4.29LDD109 pKa = 3.95NASFTLYY116 pKa = 10.73DD117 pKa = 3.53YY118 pKa = 11.67LLYY121 pKa = 10.9DD122 pKa = 3.53NQAYY126 pKa = 10.48LSDD129 pKa = 5.4AINEE133 pKa = 4.24VADD136 pKa = 3.64QSVDD140 pKa = 2.68IYY142 pKa = 11.6YY143 pKa = 10.66SDD145 pKa = 4.45LLKK148 pKa = 10.86SLPDD152 pKa = 3.8LEE154 pKa = 4.91NGGYY158 pKa = 7.84YY159 pKa = 9.86EE160 pKa = 4.11RR161 pKa = 11.84AAEE164 pKa = 4.08EE165 pKa = 4.08FGLTGDD171 pKa = 4.45LYY173 pKa = 11.27KK174 pKa = 10.92DD175 pKa = 3.07IMTAQYY181 pKa = 10.68LRR183 pKa = 11.84NIDD186 pKa = 3.82EE187 pKa = 4.95LYY189 pKa = 11.02EE190 pKa = 4.15NLDD193 pKa = 4.59DD194 pKa = 3.96IVKK197 pKa = 10.46SVAISIAEE205 pKa = 4.23DD206 pKa = 3.66YY207 pKa = 9.54EE208 pKa = 4.56LPLTEE213 pKa = 5.38GIEE216 pKa = 3.96EE217 pKa = 4.19ALNNISDD224 pKa = 4.36CNKK227 pKa = 9.33HH228 pKa = 6.85DD229 pKa = 4.73SIEE232 pKa = 4.34PIIDD236 pKa = 3.61EE237 pKa = 4.56LKK239 pKa = 10.29DD240 pKa = 3.57TIGYY244 pKa = 7.49EE245 pKa = 3.94AQDD248 pKa = 3.19IAKK251 pKa = 9.36EE252 pKa = 3.99KK253 pKa = 10.55LSRR256 pKa = 11.84EE257 pKa = 4.37GIDD260 pKa = 3.11INDD263 pKa = 4.89DD264 pKa = 3.13IEE266 pKa = 4.37EE267 pKa = 4.48YY268 pKa = 10.33IEE270 pKa = 4.69EE271 pKa = 4.09ITVYY275 pKa = 10.55EE276 pKa = 4.01LDD278 pKa = 3.96GNQNMMDD285 pKa = 5.15LINDD289 pKa = 3.97KK290 pKa = 10.73IEE292 pKa = 4.41EE293 pKa = 4.13IKK295 pKa = 10.76EE296 pKa = 3.72DD297 pKa = 3.87LEE299 pKa = 4.39INKK302 pKa = 10.06DD303 pKa = 3.4MSRR306 pKa = 3.42
MM1 pKa = 7.56SYY3 pKa = 11.03YY4 pKa = 10.25EE5 pKa = 3.7VRR7 pKa = 11.84ANVNQLISEE16 pKa = 4.18NYY18 pKa = 7.77EE19 pKa = 4.08DD20 pKa = 4.18YY21 pKa = 11.09VKK23 pKa = 11.07SLISIEE29 pKa = 4.21KK30 pKa = 10.03NINNEE35 pKa = 3.77DD36 pKa = 4.15CLNSIYY42 pKa = 11.06NEE44 pKa = 4.52FINDD48 pKa = 3.18NNAASIFDD56 pKa = 4.21ADD58 pKa = 3.04IDD60 pKa = 4.42GYY62 pKa = 10.22IEE64 pKa = 4.13KK65 pKa = 10.62LGIDD69 pKa = 3.94LEE71 pKa = 4.46DD72 pKa = 3.35EE73 pKa = 4.27RR74 pKa = 11.84VIHH77 pKa = 6.32GEE79 pKa = 3.71PVSYY83 pKa = 10.81EE84 pKa = 3.91EE85 pKa = 4.01EE86 pKa = 4.0QEE88 pKa = 4.31RR89 pKa = 11.84EE90 pKa = 3.51VDD92 pKa = 3.25RR93 pKa = 11.84EE94 pKa = 4.03KK95 pKa = 11.11FNKK98 pKa = 9.52NDD100 pKa = 3.63YY101 pKa = 10.86EE102 pKa = 4.15NLKK105 pKa = 10.14PDD107 pKa = 4.29LDD109 pKa = 3.95NASFTLYY116 pKa = 10.73DD117 pKa = 3.53YY118 pKa = 11.67LLYY121 pKa = 10.9DD122 pKa = 3.53NQAYY126 pKa = 10.48LSDD129 pKa = 5.4AINEE133 pKa = 4.24VADD136 pKa = 3.64QSVDD140 pKa = 2.68IYY142 pKa = 11.6YY143 pKa = 10.66SDD145 pKa = 4.45LLKK148 pKa = 10.86SLPDD152 pKa = 3.8LEE154 pKa = 4.91NGGYY158 pKa = 7.84YY159 pKa = 9.86EE160 pKa = 4.11RR161 pKa = 11.84AAEE164 pKa = 4.08EE165 pKa = 4.08FGLTGDD171 pKa = 4.45LYY173 pKa = 11.27KK174 pKa = 10.92DD175 pKa = 3.07IMTAQYY181 pKa = 10.68LRR183 pKa = 11.84NIDD186 pKa = 3.82EE187 pKa = 4.95LYY189 pKa = 11.02EE190 pKa = 4.15NLDD193 pKa = 4.59DD194 pKa = 3.96IVKK197 pKa = 10.46SVAISIAEE205 pKa = 4.23DD206 pKa = 3.66YY207 pKa = 9.54EE208 pKa = 4.56LPLTEE213 pKa = 5.38GIEE216 pKa = 3.96EE217 pKa = 4.19ALNNISDD224 pKa = 4.36CNKK227 pKa = 9.33HH228 pKa = 6.85DD229 pKa = 4.73SIEE232 pKa = 4.34PIIDD236 pKa = 3.61EE237 pKa = 4.56LKK239 pKa = 10.29DD240 pKa = 3.57TIGYY244 pKa = 7.49EE245 pKa = 3.94AQDD248 pKa = 3.19IAKK251 pKa = 9.36EE252 pKa = 3.99KK253 pKa = 10.55LSRR256 pKa = 11.84EE257 pKa = 4.37GIDD260 pKa = 3.11INDD263 pKa = 4.89DD264 pKa = 3.13IEE266 pKa = 4.37EE267 pKa = 4.48YY268 pKa = 10.33IEE270 pKa = 4.69EE271 pKa = 4.09ITVYY275 pKa = 10.55EE276 pKa = 4.01LDD278 pKa = 3.96GNQNMMDD285 pKa = 5.15LINDD289 pKa = 3.97KK290 pKa = 10.73IEE292 pKa = 4.41EE293 pKa = 4.13IKK295 pKa = 10.76EE296 pKa = 3.72DD297 pKa = 3.87LEE299 pKa = 4.39INKK302 pKa = 10.06DD303 pKa = 3.4MSRR306 pKa = 3.42
Molecular weight: 35.49 kDa
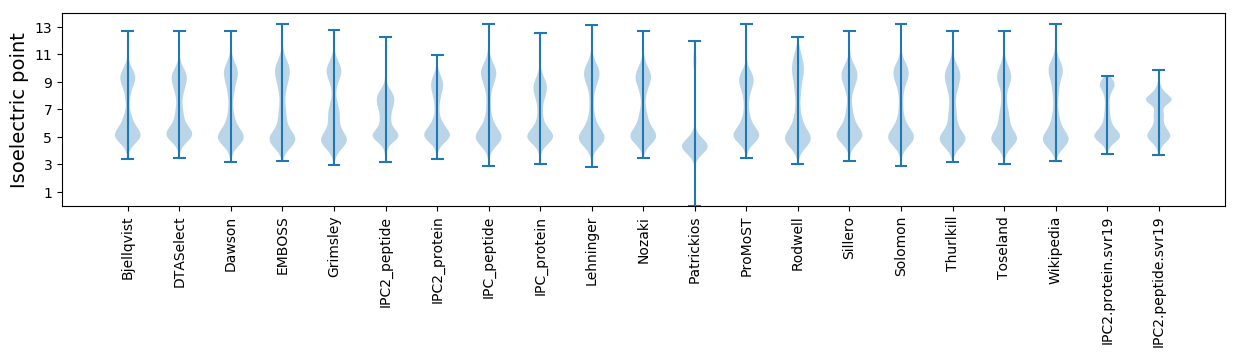
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C2BE23|C2BE23_9FIRM Glucose/Sorbosone dehydrogenase (Fragment) OS=Anaerococcus lactolyticus ATCC 51172 OX=525254 GN=HMPREF0072_0593 PE=4 SV=1
LL1 pKa = 6.4QAGVRR6 pKa = 11.84PAVRR10 pKa = 11.84QPLRR14 pKa = 11.84LPPXGAGRR22 pKa = 11.84GHH24 pKa = 6.37LQGRR28 pKa = 11.84VQALSGNAGRR38 pKa = 11.84RR39 pKa = 11.84QAVLHH44 pKa = 6.42PGRR47 pKa = 11.84CGALRR52 pKa = 11.84TVRR55 pKa = 11.84GQHH58 pKa = 6.67LLGHH62 pKa = 6.07PWWRR66 pKa = 11.84AGAGVPGVRR75 pKa = 11.84RR76 pKa = 11.84LQAAA80 pKa = 3.74
LL1 pKa = 6.4QAGVRR6 pKa = 11.84PAVRR10 pKa = 11.84QPLRR14 pKa = 11.84LPPXGAGRR22 pKa = 11.84GHH24 pKa = 6.37LQGRR28 pKa = 11.84VQALSGNAGRR38 pKa = 11.84RR39 pKa = 11.84QAVLHH44 pKa = 6.42PGRR47 pKa = 11.84CGALRR52 pKa = 11.84TVRR55 pKa = 11.84GQHH58 pKa = 6.67LLGHH62 pKa = 6.07PWWRR66 pKa = 11.84AGAGVPGVRR75 pKa = 11.84RR76 pKa = 11.84LQAAA80 pKa = 3.74
Molecular weight: 8.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
650064 |
29 |
5653 |
288.7 |
32.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.233 ± 0.055 | 0.755 ± 0.018 |
6.722 ± 0.047 | 7.55 ± 0.061 |
4.638 ± 0.043 | 6.264 ± 0.05 |
1.308 ± 0.021 | 9.154 ± 0.064 |
9.464 ± 0.071 | 9.32 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.506 ± 0.033 | 5.653 ± 0.046 |
2.75 ± 0.035 | 2.347 ± 0.028 |
3.756 ± 0.04 | 6.131 ± 0.039 |
4.7 ± 0.051 | 5.925 ± 0.047 |
0.616 ± 0.014 | 4.209 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
