
Methylobacterium symbioticum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
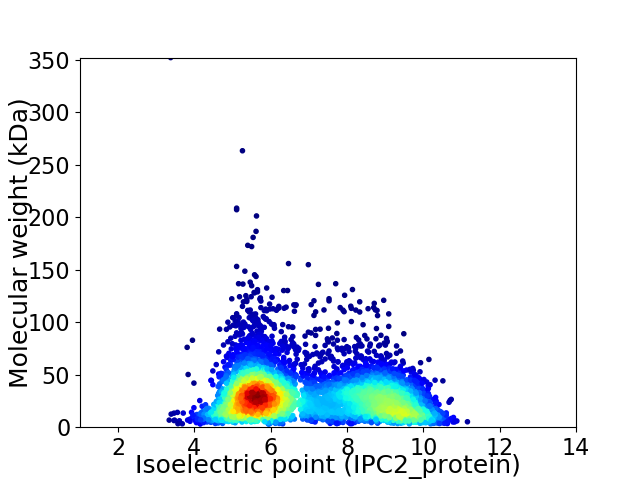
Virtual 2D-PAGE plot for 5664 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A509E7Z2|A0A509E7Z2_9RHIZ Putative glucarate transporter OS=Methylobacterium symbioticum OX=2584084 GN=gudP_1 PE=4 SV=1
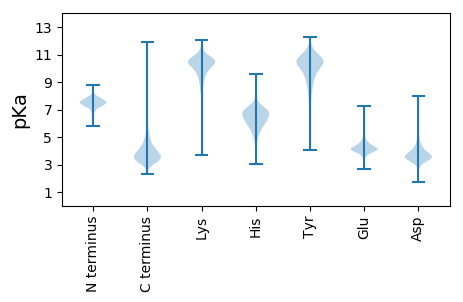
MM1 pKa = 7.6VDD3 pKa = 3.31LTGYY7 pKa = 10.93RR8 pKa = 11.84LTFDD12 pKa = 5.35DD13 pKa = 5.19EE14 pKa = 4.8FNQLSVSATGSGTVWASTRR33 pKa = 11.84PGSILRR39 pKa = 11.84PGVDD43 pKa = 2.43IGFGQSAFVDD53 pKa = 4.47PSTGVQPFQIVNGALQIQAAPATGAVADD81 pKa = 4.29LVYY84 pKa = 10.61PGLWTSGLLQASNSFSQTYY103 pKa = 10.29GYY105 pKa = 11.13FEE107 pKa = 4.19MRR109 pKa = 11.84AEE111 pKa = 4.33LPAEE115 pKa = 4.16TGAWPGFWLLNADD128 pKa = 3.95GVWPPEE134 pKa = 3.59IDD136 pKa = 3.08IVEE139 pKa = 4.56AYY141 pKa = 10.24GADD144 pKa = 3.47PTALTNTIHH153 pKa = 6.19TGEE156 pKa = 4.34TGQPTHH162 pKa = 4.83QTIWSNQPTLSAGYY176 pKa = 8.44HH177 pKa = 4.96TFGALWTASTITFFYY192 pKa = 10.62DD193 pKa = 3.64GNPVGQVATPSDD205 pKa = 3.91MHH207 pKa = 8.0GPMYY211 pKa = 10.39PLVNLAITRR220 pKa = 11.84GIEE223 pKa = 4.38GITDD227 pKa = 3.62SVKK230 pKa = 10.13TMSVDD235 pKa = 3.47YY236 pKa = 10.71IRR238 pKa = 11.84VYY240 pKa = 10.87SADD243 pKa = 3.53PSATAVALQTVSSPDD258 pKa = 3.43GADD261 pKa = 2.83TGGLYY266 pKa = 10.37GASASNVTPPPPTAKK281 pKa = 10.06PDD283 pKa = 3.48TLTIRR288 pKa = 11.84VSGDD292 pKa = 2.65SWNGAPKK299 pKa = 10.2FVVSVDD305 pKa = 3.59GTPLGQVFSVDD316 pKa = 3.2ADD318 pKa = 3.65HH319 pKa = 7.62AAGQWQDD326 pKa = 3.07VTVQGNFGAGPHH338 pKa = 5.05QVAVTYY344 pKa = 10.82INDD347 pKa = 3.7GNEE350 pKa = 3.74NPYY353 pKa = 10.79YY354 pKa = 10.53DD355 pKa = 3.62AAANKK360 pKa = 9.33VFNDD364 pKa = 3.05RR365 pKa = 11.84NLFVTGIALNGTSYY379 pKa = 11.83GLGSVVSNSASVGWDD394 pKa = 3.22LLDD397 pKa = 4.19PNAAVMAANGTLIFGTAGGTVTPIDD422 pKa = 4.43PPTTPALPTTPATPTTPTAPAAGQDD447 pKa = 3.39TLTIRR452 pKa = 11.84ISGDD456 pKa = 2.77SWNGAPQFVVSVDD469 pKa = 3.46GTQVAGPFDD478 pKa = 3.79VTADD482 pKa = 3.47HH483 pKa = 7.46AAGQWQDD490 pKa = 3.09VSVQGDD496 pKa = 4.34FGAGPHH502 pKa = 5.35QVTITYY508 pKa = 10.67LNDD511 pKa = 3.41GNEE514 pKa = 4.1NPYY517 pKa = 10.71YY518 pKa = 10.53DD519 pKa = 3.94AVANKK524 pKa = 9.45VFNDD528 pKa = 3.13RR529 pKa = 11.84NLFVTGIALNGTAYY543 pKa = 10.55GLDD546 pKa = 3.03AVVSNAAAAGWDD558 pKa = 3.5RR559 pKa = 11.84LDD561 pKa = 3.85PNAAVMAASGTLVFGTGAATPGTSSPTTPLPTTPTAPVTAPGQDD605 pKa = 3.04TLTIHH610 pKa = 7.33ISGDD614 pKa = 3.15SWNGAPQFVVSVDD627 pKa = 3.44GTSVGGPFDD636 pKa = 3.46VTADD640 pKa = 3.47HH641 pKa = 7.46AAGQWQDD648 pKa = 3.09VSVQGDD654 pKa = 4.34FGAGPHH660 pKa = 5.35QVTITYY666 pKa = 10.67LNDD669 pKa = 3.41GNEE672 pKa = 4.1NPYY675 pKa = 10.71YY676 pKa = 10.53DD677 pKa = 3.94AVANKK682 pKa = 9.45VFNDD686 pKa = 3.13RR687 pKa = 11.84NLFVTGIDD695 pKa = 3.99LNGTAYY701 pKa = 10.53GLDD704 pKa = 2.97AVVSNMAAAGWDD716 pKa = 3.44RR717 pKa = 11.84LDD719 pKa = 3.85PNAAVMAASGTLVFGTGAA737 pKa = 3.44
MM1 pKa = 7.6VDD3 pKa = 3.31LTGYY7 pKa = 10.93RR8 pKa = 11.84LTFDD12 pKa = 5.35DD13 pKa = 5.19EE14 pKa = 4.8FNQLSVSATGSGTVWASTRR33 pKa = 11.84PGSILRR39 pKa = 11.84PGVDD43 pKa = 2.43IGFGQSAFVDD53 pKa = 4.47PSTGVQPFQIVNGALQIQAAPATGAVADD81 pKa = 4.29LVYY84 pKa = 10.61PGLWTSGLLQASNSFSQTYY103 pKa = 10.29GYY105 pKa = 11.13FEE107 pKa = 4.19MRR109 pKa = 11.84AEE111 pKa = 4.33LPAEE115 pKa = 4.16TGAWPGFWLLNADD128 pKa = 3.95GVWPPEE134 pKa = 3.59IDD136 pKa = 3.08IVEE139 pKa = 4.56AYY141 pKa = 10.24GADD144 pKa = 3.47PTALTNTIHH153 pKa = 6.19TGEE156 pKa = 4.34TGQPTHH162 pKa = 4.83QTIWSNQPTLSAGYY176 pKa = 8.44HH177 pKa = 4.96TFGALWTASTITFFYY192 pKa = 10.62DD193 pKa = 3.64GNPVGQVATPSDD205 pKa = 3.91MHH207 pKa = 8.0GPMYY211 pKa = 10.39PLVNLAITRR220 pKa = 11.84GIEE223 pKa = 4.38GITDD227 pKa = 3.62SVKK230 pKa = 10.13TMSVDD235 pKa = 3.47YY236 pKa = 10.71IRR238 pKa = 11.84VYY240 pKa = 10.87SADD243 pKa = 3.53PSATAVALQTVSSPDD258 pKa = 3.43GADD261 pKa = 2.83TGGLYY266 pKa = 10.37GASASNVTPPPPTAKK281 pKa = 10.06PDD283 pKa = 3.48TLTIRR288 pKa = 11.84VSGDD292 pKa = 2.65SWNGAPKK299 pKa = 10.2FVVSVDD305 pKa = 3.59GTPLGQVFSVDD316 pKa = 3.2ADD318 pKa = 3.65HH319 pKa = 7.62AAGQWQDD326 pKa = 3.07VTVQGNFGAGPHH338 pKa = 5.05QVAVTYY344 pKa = 10.82INDD347 pKa = 3.7GNEE350 pKa = 3.74NPYY353 pKa = 10.79YY354 pKa = 10.53DD355 pKa = 3.62AAANKK360 pKa = 9.33VFNDD364 pKa = 3.05RR365 pKa = 11.84NLFVTGIALNGTSYY379 pKa = 11.83GLGSVVSNSASVGWDD394 pKa = 3.22LLDD397 pKa = 4.19PNAAVMAANGTLIFGTAGGTVTPIDD422 pKa = 4.43PPTTPALPTTPATPTTPTAPAAGQDD447 pKa = 3.39TLTIRR452 pKa = 11.84ISGDD456 pKa = 2.77SWNGAPQFVVSVDD469 pKa = 3.46GTQVAGPFDD478 pKa = 3.79VTADD482 pKa = 3.47HH483 pKa = 7.46AAGQWQDD490 pKa = 3.09VSVQGDD496 pKa = 4.34FGAGPHH502 pKa = 5.35QVTITYY508 pKa = 10.67LNDD511 pKa = 3.41GNEE514 pKa = 4.1NPYY517 pKa = 10.71YY518 pKa = 10.53DD519 pKa = 3.94AVANKK524 pKa = 9.45VFNDD528 pKa = 3.13RR529 pKa = 11.84NLFVTGIALNGTAYY543 pKa = 10.55GLDD546 pKa = 3.03AVVSNAAAAGWDD558 pKa = 3.5RR559 pKa = 11.84LDD561 pKa = 3.85PNAAVMAASGTLVFGTGAATPGTSSPTTPLPTTPTAPVTAPGQDD605 pKa = 3.04TLTIHH610 pKa = 7.33ISGDD614 pKa = 3.15SWNGAPQFVVSVDD627 pKa = 3.44GTSVGGPFDD636 pKa = 3.46VTADD640 pKa = 3.47HH641 pKa = 7.46AAGQWQDD648 pKa = 3.09VSVQGDD654 pKa = 4.34FGAGPHH660 pKa = 5.35QVTITYY666 pKa = 10.67LNDD669 pKa = 3.41GNEE672 pKa = 4.1NPYY675 pKa = 10.71YY676 pKa = 10.53DD677 pKa = 3.94AVANKK682 pKa = 9.45VFNDD686 pKa = 3.13RR687 pKa = 11.84NLFVTGIDD695 pKa = 3.99LNGTAYY701 pKa = 10.53GLDD704 pKa = 2.97AVVSNMAAAGWDD716 pKa = 3.44RR717 pKa = 11.84LDD719 pKa = 3.85PNAAVMAASGTLVFGTGAA737 pKa = 3.44
Molecular weight: 76.04 kDa
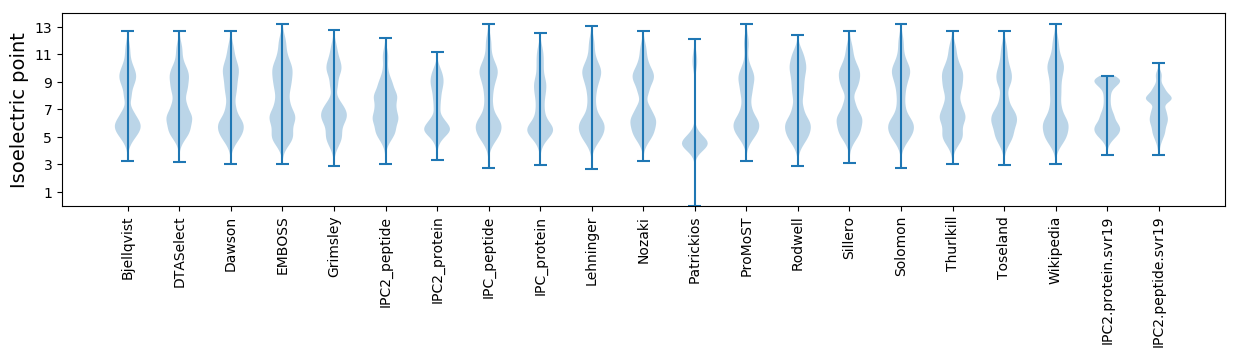
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A509EDV8|A0A509EDV8_9RHIZ Lead cadmium zinc and mercury-transporting ATPase OS=Methylobacterium symbioticum OX=2584084 GN=zntA_3 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.12VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.95GRR39 pKa = 11.84KK40 pKa = 9.25RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.12VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.95GRR39 pKa = 11.84KK40 pKa = 9.25RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1690294 |
29 |
3678 |
298.4 |
32.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.447 ± 0.059 | 0.845 ± 0.011 |
5.403 ± 0.024 | 5.516 ± 0.03 |
3.329 ± 0.021 | 9.209 ± 0.043 |
1.944 ± 0.014 | 4.349 ± 0.025 |
2.415 ± 0.03 | 10.621 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.044 ± 0.016 | 2.051 ± 0.019 |
5.847 ± 0.029 | 2.852 ± 0.021 |
8.285 ± 0.042 | 4.81 ± 0.025 |
5.213 ± 0.032 | 7.584 ± 0.027 |
1.219 ± 0.014 | 2.018 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
