
Shigella phage SfII (Shigella flexneri bacteriophage II) (Bacteriophage SfII)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
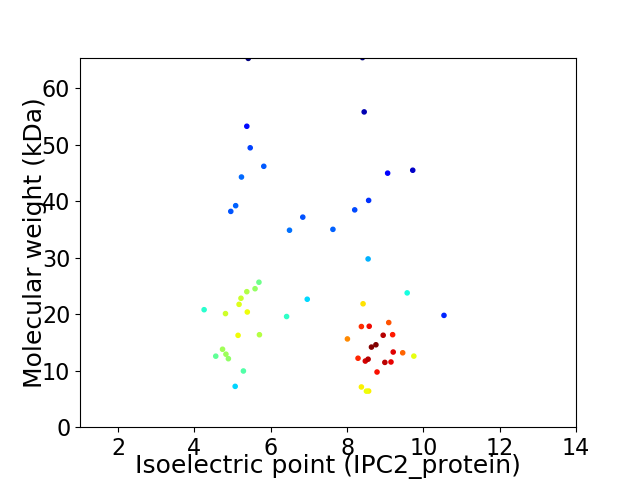
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5FM63|S5FM63_BPSF2 Probable tape measure protein OS=Shigella phage SfII OX=66284 GN=SfII_16 PE=3 SV=1
MM1 pKa = 7.69KK2 pKa = 8.86LTPVIAALRR11 pKa = 11.84ARR13 pKa = 11.84CPYY16 pKa = 10.06FEE18 pKa = 4.39NRR20 pKa = 11.84VAGAAQFKK28 pKa = 10.19NLPEE32 pKa = 4.11VGKK35 pKa = 10.51LRR37 pKa = 11.84LPAAYY42 pKa = 9.22VVPGDD47 pKa = 4.84DD48 pKa = 4.34SPGEE52 pKa = 4.13NKK54 pKa = 10.35SQTDD58 pKa = 3.3YY59 pKa = 10.55WQEE62 pKa = 3.75LKK64 pKa = 10.91EE65 pKa = 4.23GFSVVVILSNGRR77 pKa = 11.84DD78 pKa = 3.43EE79 pKa = 5.21RR80 pKa = 11.84GQFASYY86 pKa = 11.14DD87 pKa = 3.57VVDD90 pKa = 5.36DD91 pKa = 3.79VRR93 pKa = 11.84QMLFKK98 pKa = 10.84ALLGWNPEE106 pKa = 3.8ACGNPITYY114 pKa = 10.35DD115 pKa = 3.18GGTLLDD121 pKa = 4.22LNRR124 pKa = 11.84HH125 pKa = 4.61EE126 pKa = 6.12LIYY129 pKa = 10.88QFDD132 pKa = 3.96FSVISEE138 pKa = 4.21LTEE141 pKa = 4.82DD142 pKa = 4.41DD143 pKa = 3.88TRR145 pKa = 11.84QQDD148 pKa = 3.89DD149 pKa = 4.49LNSLDD154 pKa = 4.01EE155 pKa = 4.63LRR157 pKa = 11.84TLAIDD162 pKa = 4.15VDD164 pKa = 4.29YY165 pKa = 10.82LDD167 pKa = 5.29PGNGPDD173 pKa = 4.82GDD175 pKa = 4.3IEE177 pKa = 4.36HH178 pKa = 6.63HH179 pKa = 5.92TEE181 pKa = 3.68IPLPSS186 pKa = 3.38
MM1 pKa = 7.69KK2 pKa = 8.86LTPVIAALRR11 pKa = 11.84ARR13 pKa = 11.84CPYY16 pKa = 10.06FEE18 pKa = 4.39NRR20 pKa = 11.84VAGAAQFKK28 pKa = 10.19NLPEE32 pKa = 4.11VGKK35 pKa = 10.51LRR37 pKa = 11.84LPAAYY42 pKa = 9.22VVPGDD47 pKa = 4.84DD48 pKa = 4.34SPGEE52 pKa = 4.13NKK54 pKa = 10.35SQTDD58 pKa = 3.3YY59 pKa = 10.55WQEE62 pKa = 3.75LKK64 pKa = 10.91EE65 pKa = 4.23GFSVVVILSNGRR77 pKa = 11.84DD78 pKa = 3.43EE79 pKa = 5.21RR80 pKa = 11.84GQFASYY86 pKa = 11.14DD87 pKa = 3.57VVDD90 pKa = 5.36DD91 pKa = 3.79VRR93 pKa = 11.84QMLFKK98 pKa = 10.84ALLGWNPEE106 pKa = 3.8ACGNPITYY114 pKa = 10.35DD115 pKa = 3.18GGTLLDD121 pKa = 4.22LNRR124 pKa = 11.84HH125 pKa = 4.61EE126 pKa = 6.12LIYY129 pKa = 10.88QFDD132 pKa = 3.96FSVISEE138 pKa = 4.21LTEE141 pKa = 4.82DD142 pKa = 4.41DD143 pKa = 3.88TRR145 pKa = 11.84QQDD148 pKa = 3.89DD149 pKa = 4.49LNSLDD154 pKa = 4.01EE155 pKa = 4.63LRR157 pKa = 11.84TLAIDD162 pKa = 4.15VDD164 pKa = 4.29YY165 pKa = 10.82LDD167 pKa = 5.29PGNGPDD173 pKa = 4.82GDD175 pKa = 4.3IEE177 pKa = 4.36HH178 pKa = 6.63HH179 pKa = 5.92TEE181 pKa = 3.68IPLPSS186 pKa = 3.38
Molecular weight: 20.79 kDa
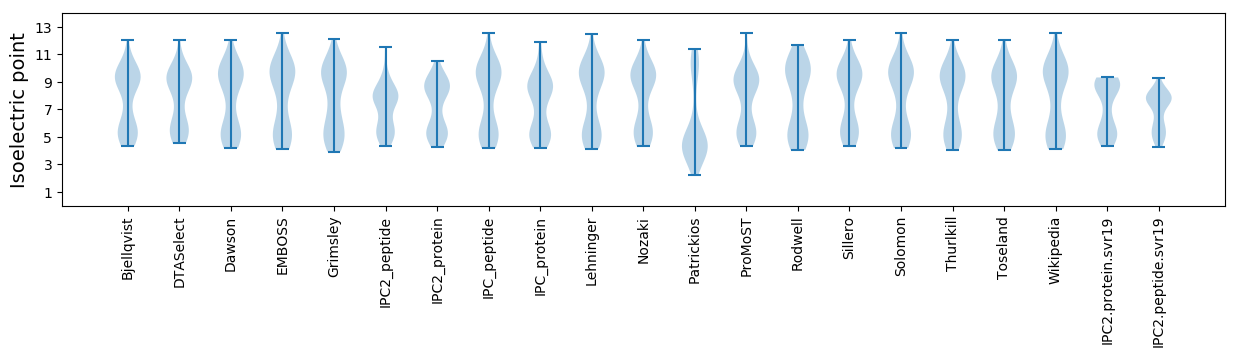
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5FM74|S5FM74_BPSF2 Excisionase OS=Shigella phage SfII OX=66284 GN=xis PE=4 SV=1
MM1 pKa = 7.9PARR4 pKa = 11.84KK5 pKa = 9.27VCQTFFRR12 pKa = 11.84NALASTHH19 pKa = 5.94QYY21 pKa = 8.87RR22 pKa = 11.84QNAIIDD28 pKa = 3.71SAAALVGGASLSLTSIGRR46 pKa = 11.84HH47 pKa = 4.94LPGPARR53 pKa = 11.84VKK55 pKa = 10.94DD56 pKa = 3.56KK57 pKa = 10.77IKK59 pKa = 10.81RR60 pKa = 11.84VDD62 pKa = 3.25RR63 pKa = 11.84LLGNKK68 pKa = 9.15RR69 pKa = 11.84LHH71 pKa = 6.51NDD73 pKa = 2.37IPLIFKK79 pKa = 10.59NITSMMTNKK88 pKa = 10.3LSWCVIAVDD97 pKa = 3.42WSGYY101 pKa = 9.02PFQEE105 pKa = 4.02YY106 pKa = 9.92HH107 pKa = 5.94VLRR110 pKa = 11.84ASLLCDD116 pKa = 3.31GRR118 pKa = 11.84SIPLMSQVFPSKK130 pKa = 10.69KK131 pKa = 9.45NNEE134 pKa = 4.0AVEE137 pKa = 4.15IAFLDD142 pKa = 4.01ALSGAISPRR151 pKa = 11.84TRR153 pKa = 11.84VVIVTDD159 pKa = 4.57AGFQSAWFRR168 pKa = 11.84HH169 pKa = 5.7IKK171 pKa = 8.66SQGWDD176 pKa = 3.72FIGRR180 pKa = 11.84IRR182 pKa = 11.84GVVKK186 pKa = 10.52FRR188 pKa = 11.84LDD190 pKa = 3.24SDD192 pKa = 3.48KK193 pKa = 11.59DD194 pKa = 3.04KK195 pKa = 11.08WLDD198 pKa = 3.08IKK200 pKa = 10.03MCRR203 pKa = 11.84GSSEE207 pKa = 4.03AKK209 pKa = 9.91YY210 pKa = 11.05LEE212 pKa = 4.52TGTLARR218 pKa = 11.84KK219 pKa = 9.34KK220 pKa = 9.96RR221 pKa = 11.84SQCEE225 pKa = 3.68GHH227 pKa = 6.37FYY229 pKa = 10.7LYY231 pKa = 10.48KK232 pKa = 10.52HH233 pKa = 5.87SPKK236 pKa = 10.08GRR238 pKa = 11.84KK239 pKa = 7.75SRR241 pKa = 11.84RR242 pKa = 11.84ARR244 pKa = 11.84GRR246 pKa = 11.84PGLPTTEE253 pKa = 4.73KK254 pKa = 9.27EE255 pKa = 3.8QKK257 pKa = 10.11AAGRR261 pKa = 11.84EE262 pKa = 3.69PWLIFSNTAEE272 pKa = 4.16FNAKK276 pKa = 10.04KK277 pKa = 10.2IMKK280 pKa = 9.46LYY282 pKa = 9.4SRR284 pKa = 11.84RR285 pKa = 11.84MQIEE289 pKa = 3.85QNFRR293 pKa = 11.84DD294 pKa = 3.9EE295 pKa = 3.93KK296 pKa = 10.64SEE298 pKa = 3.97RR299 pKa = 11.84FGFGLRR305 pKa = 11.84ASRR308 pKa = 11.84SRR310 pKa = 11.84RR311 pKa = 11.84GEE313 pKa = 3.82RR314 pKa = 11.84FLVLSLLVTLASIVLWLLGYY334 pKa = 10.4HH335 pKa = 6.12FEE337 pKa = 4.76NKK339 pKa = 9.56GFHH342 pKa = 6.7LKK344 pKa = 10.04YY345 pKa = 9.97QANSLKK351 pKa = 10.15KK352 pKa = 9.98RR353 pKa = 11.84RR354 pKa = 11.84VLSFLTLAEE363 pKa = 3.95NVLRR367 pKa = 11.84FNPEE371 pKa = 3.79LLRR374 pKa = 11.84RR375 pKa = 11.84AQPEE379 pKa = 4.21KK380 pKa = 10.34ILCRR384 pKa = 11.84LASTYY389 pKa = 10.96RR390 pKa = 11.84SMVLAYY396 pKa = 10.23
MM1 pKa = 7.9PARR4 pKa = 11.84KK5 pKa = 9.27VCQTFFRR12 pKa = 11.84NALASTHH19 pKa = 5.94QYY21 pKa = 8.87RR22 pKa = 11.84QNAIIDD28 pKa = 3.71SAAALVGGASLSLTSIGRR46 pKa = 11.84HH47 pKa = 4.94LPGPARR53 pKa = 11.84VKK55 pKa = 10.94DD56 pKa = 3.56KK57 pKa = 10.77IKK59 pKa = 10.81RR60 pKa = 11.84VDD62 pKa = 3.25RR63 pKa = 11.84LLGNKK68 pKa = 9.15RR69 pKa = 11.84LHH71 pKa = 6.51NDD73 pKa = 2.37IPLIFKK79 pKa = 10.59NITSMMTNKK88 pKa = 10.3LSWCVIAVDD97 pKa = 3.42WSGYY101 pKa = 9.02PFQEE105 pKa = 4.02YY106 pKa = 9.92HH107 pKa = 5.94VLRR110 pKa = 11.84ASLLCDD116 pKa = 3.31GRR118 pKa = 11.84SIPLMSQVFPSKK130 pKa = 10.69KK131 pKa = 9.45NNEE134 pKa = 4.0AVEE137 pKa = 4.15IAFLDD142 pKa = 4.01ALSGAISPRR151 pKa = 11.84TRR153 pKa = 11.84VVIVTDD159 pKa = 4.57AGFQSAWFRR168 pKa = 11.84HH169 pKa = 5.7IKK171 pKa = 8.66SQGWDD176 pKa = 3.72FIGRR180 pKa = 11.84IRR182 pKa = 11.84GVVKK186 pKa = 10.52FRR188 pKa = 11.84LDD190 pKa = 3.24SDD192 pKa = 3.48KK193 pKa = 11.59DD194 pKa = 3.04KK195 pKa = 11.08WLDD198 pKa = 3.08IKK200 pKa = 10.03MCRR203 pKa = 11.84GSSEE207 pKa = 4.03AKK209 pKa = 9.91YY210 pKa = 11.05LEE212 pKa = 4.52TGTLARR218 pKa = 11.84KK219 pKa = 9.34KK220 pKa = 9.96RR221 pKa = 11.84SQCEE225 pKa = 3.68GHH227 pKa = 6.37FYY229 pKa = 10.7LYY231 pKa = 10.48KK232 pKa = 10.52HH233 pKa = 5.87SPKK236 pKa = 10.08GRR238 pKa = 11.84KK239 pKa = 7.75SRR241 pKa = 11.84RR242 pKa = 11.84ARR244 pKa = 11.84GRR246 pKa = 11.84PGLPTTEE253 pKa = 4.73KK254 pKa = 9.27EE255 pKa = 3.8QKK257 pKa = 10.11AAGRR261 pKa = 11.84EE262 pKa = 3.69PWLIFSNTAEE272 pKa = 4.16FNAKK276 pKa = 10.04KK277 pKa = 10.2IMKK280 pKa = 9.46LYY282 pKa = 9.4SRR284 pKa = 11.84RR285 pKa = 11.84MQIEE289 pKa = 3.85QNFRR293 pKa = 11.84DD294 pKa = 3.9EE295 pKa = 3.93KK296 pKa = 10.64SEE298 pKa = 3.97RR299 pKa = 11.84FGFGLRR305 pKa = 11.84ASRR308 pKa = 11.84SRR310 pKa = 11.84RR311 pKa = 11.84GEE313 pKa = 3.82RR314 pKa = 11.84FLVLSLLVTLASIVLWLLGYY334 pKa = 10.4HH335 pKa = 6.12FEE337 pKa = 4.76NKK339 pKa = 9.56GFHH342 pKa = 6.7LKK344 pKa = 10.04YY345 pKa = 9.97QANSLKK351 pKa = 10.15KK352 pKa = 9.98RR353 pKa = 11.84RR354 pKa = 11.84VLSFLTLAEE363 pKa = 3.95NVLRR367 pKa = 11.84FNPEE371 pKa = 3.79LLRR374 pKa = 11.84RR375 pKa = 11.84AQPEE379 pKa = 4.21KK380 pKa = 10.34ILCRR384 pKa = 11.84LASTYY389 pKa = 10.96RR390 pKa = 11.84SMVLAYY396 pKa = 10.23
Molecular weight: 45.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12616 |
56 |
611 |
217.5 |
24.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.743 ± 0.354 | 1.3 ± 0.167 |
5.485 ± 0.273 | 5.588 ± 0.296 |
4.011 ± 0.294 | 6.983 ± 0.331 |
1.839 ± 0.149 | 5.929 ± 0.538 |
5.652 ± 0.325 | 9.028 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.663 ± 0.172 | 4.328 ± 0.248 |
4.574 ± 0.205 | 3.686 ± 0.199 |
6.294 ± 0.43 | 6.476 ± 0.256 |
5.77 ± 0.305 | 6.714 ± 0.262 |
1.807 ± 0.138 | 3.131 ± 0.219 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
