
Human papillomavirus 204
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Mupapillomavirus; Mupapillomavirus 3
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
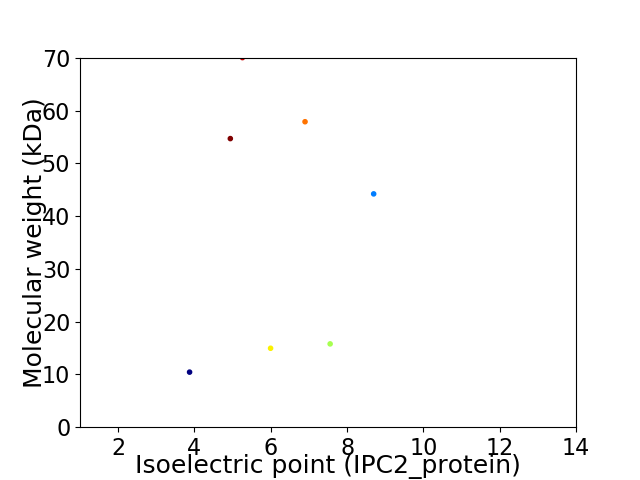
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
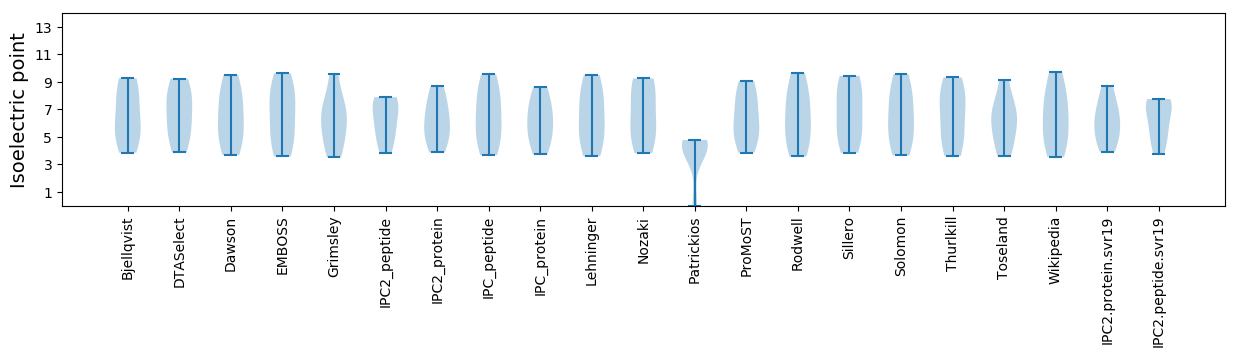
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7GGY8|A0A0F7GGY8_9PAPI Protein E6 OS=Human papillomavirus 204 OX=1650736 GN=E6 PE=3 SV=1
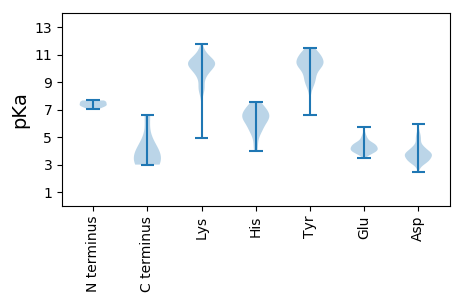
MM1 pKa = 7.26RR2 pKa = 11.84QEE4 pKa = 3.66NLAARR9 pKa = 11.84LEE11 pKa = 4.51TILLEE16 pKa = 4.17EE17 pKa = 4.6EE18 pKa = 4.42PNVLDD23 pKa = 3.26LHH25 pKa = 7.05CYY27 pKa = 10.37EE28 pKa = 4.83EE29 pKa = 4.46VALSDD34 pKa = 4.02EE35 pKa = 4.63EE36 pKa = 5.09EE37 pKa = 4.38EE38 pKa = 4.41SQQVQQPYY46 pKa = 10.58LVTVPCAQCLNVVTFTCAADD66 pKa = 3.69LLSIRR71 pKa = 11.84EE72 pKa = 4.08LQQLLFDD79 pKa = 4.78CLFLCEE85 pKa = 3.96TCAAEE90 pKa = 4.93LSS92 pKa = 3.94
MM1 pKa = 7.26RR2 pKa = 11.84QEE4 pKa = 3.66NLAARR9 pKa = 11.84LEE11 pKa = 4.51TILLEE16 pKa = 4.17EE17 pKa = 4.6EE18 pKa = 4.42PNVLDD23 pKa = 3.26LHH25 pKa = 7.05CYY27 pKa = 10.37EE28 pKa = 4.83EE29 pKa = 4.46VALSDD34 pKa = 4.02EE35 pKa = 4.63EE36 pKa = 5.09EE37 pKa = 4.38EE38 pKa = 4.41SQQVQQPYY46 pKa = 10.58LVTVPCAQCLNVVTFTCAADD66 pKa = 3.69LLSIRR71 pKa = 11.84EE72 pKa = 4.08LQQLLFDD79 pKa = 4.78CLFLCEE85 pKa = 3.96TCAAEE90 pKa = 4.93LSS92 pKa = 3.94
Molecular weight: 10.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7GG55|A0A0F7GG55_9PAPI Protein E7 OS=Human papillomavirus 204 OX=1650736 GN=E7 PE=3 SV=1
MM1 pKa = 7.66EE2 pKa = 5.39NLSQRR7 pKa = 11.84LEE9 pKa = 4.17SLQEE13 pKa = 3.95RR14 pKa = 11.84LLSLYY19 pKa = 10.04EE20 pKa = 3.82QDD22 pKa = 4.89SNDD25 pKa = 3.17IQDD28 pKa = 5.09QITHH32 pKa = 5.25WTLIKK37 pKa = 10.21QEE39 pKa = 4.09QVLFHH44 pKa = 6.01YY45 pKa = 10.33ARR47 pKa = 11.84QNGVRR52 pKa = 11.84RR53 pKa = 11.84LGMQTIPTLAASEE66 pKa = 4.24ARR68 pKa = 11.84AKK70 pKa = 10.04QAIEE74 pKa = 4.12MVLQLQSLANSPFGMEE90 pKa = 3.79PWLLQDD96 pKa = 3.74TSRR99 pKa = 11.84EE100 pKa = 4.08RR101 pKa = 11.84YY102 pKa = 6.59TAAPGNTFKK111 pKa = 10.94KK112 pKa = 10.29QPQTLLLTFDD122 pKa = 3.7NDD124 pKa = 3.14KK125 pKa = 11.3DD126 pKa = 3.76NSVEE130 pKa = 4.19HH131 pKa = 5.74TVWTYY136 pKa = 10.82IYY138 pKa = 8.74YY139 pKa = 10.59QNGDD143 pKa = 4.2GIWHH147 pKa = 6.49KK148 pKa = 10.74EE149 pKa = 3.7EE150 pKa = 4.53SGVDD154 pKa = 3.16EE155 pKa = 5.08KK156 pKa = 11.73GIFFIKK162 pKa = 10.65YY163 pKa = 8.04GVEE166 pKa = 3.71KK167 pKa = 9.9IYY169 pKa = 10.86YY170 pKa = 10.26LSFADD175 pKa = 3.39EE176 pKa = 4.0ATRR179 pKa = 11.84YY180 pKa = 8.79SRR182 pKa = 11.84KK183 pKa = 10.22GEE185 pKa = 3.96YY186 pKa = 8.74TVHH189 pKa = 6.87FKK191 pKa = 10.0SQRR194 pKa = 11.84HH195 pKa = 5.15SYY197 pKa = 9.41NVSSVSSTSGSPGSPDD213 pKa = 3.05STEE216 pKa = 4.18TNPSSSHH223 pKa = 4.05TRR225 pKa = 11.84GAEE228 pKa = 3.86EE229 pKa = 4.28EE230 pKa = 4.43SPEE233 pKa = 4.01RR234 pKa = 11.84PVRR237 pKa = 11.84SRR239 pKa = 11.84AYY241 pKa = 8.99GQRR244 pKa = 11.84PSTSPRR250 pKa = 11.84VSSRR254 pKa = 11.84RR255 pKa = 11.84GGEE258 pKa = 3.7QGKK261 pKa = 10.16SGTGTDD267 pKa = 3.87SDD269 pKa = 4.32SGLAPPSPGDD279 pKa = 3.35VGSRR283 pKa = 11.84TTQPARR289 pKa = 11.84RR290 pKa = 11.84NQSRR294 pKa = 11.84LRR296 pKa = 11.84VLLQEE301 pKa = 4.48ARR303 pKa = 11.84DD304 pKa = 3.86PLVLCLKK311 pKa = 10.45GGPNQLKK318 pKa = 9.96CLRR321 pKa = 11.84YY322 pKa = 8.85RR323 pKa = 11.84LKK325 pKa = 10.55KK326 pKa = 9.25QHH328 pKa = 6.59HH329 pKa = 6.29KK330 pKa = 10.8LFTKK334 pKa = 10.47ISTTWHH340 pKa = 5.59WVDD343 pKa = 3.09NTSTNRR349 pKa = 11.84VGNARR354 pKa = 11.84MLIQFLTEE362 pKa = 3.57EE363 pKa = 4.24QRR365 pKa = 11.84NHH367 pKa = 6.35FLDD370 pKa = 4.37VIIVPKK376 pKa = 10.31DD377 pKa = 2.89ISVYY381 pKa = 9.83RR382 pKa = 11.84GYY384 pKa = 11.15FKK386 pKa = 11.26GFF388 pKa = 3.26
MM1 pKa = 7.66EE2 pKa = 5.39NLSQRR7 pKa = 11.84LEE9 pKa = 4.17SLQEE13 pKa = 3.95RR14 pKa = 11.84LLSLYY19 pKa = 10.04EE20 pKa = 3.82QDD22 pKa = 4.89SNDD25 pKa = 3.17IQDD28 pKa = 5.09QITHH32 pKa = 5.25WTLIKK37 pKa = 10.21QEE39 pKa = 4.09QVLFHH44 pKa = 6.01YY45 pKa = 10.33ARR47 pKa = 11.84QNGVRR52 pKa = 11.84RR53 pKa = 11.84LGMQTIPTLAASEE66 pKa = 4.24ARR68 pKa = 11.84AKK70 pKa = 10.04QAIEE74 pKa = 4.12MVLQLQSLANSPFGMEE90 pKa = 3.79PWLLQDD96 pKa = 3.74TSRR99 pKa = 11.84EE100 pKa = 4.08RR101 pKa = 11.84YY102 pKa = 6.59TAAPGNTFKK111 pKa = 10.94KK112 pKa = 10.29QPQTLLLTFDD122 pKa = 3.7NDD124 pKa = 3.14KK125 pKa = 11.3DD126 pKa = 3.76NSVEE130 pKa = 4.19HH131 pKa = 5.74TVWTYY136 pKa = 10.82IYY138 pKa = 8.74YY139 pKa = 10.59QNGDD143 pKa = 4.2GIWHH147 pKa = 6.49KK148 pKa = 10.74EE149 pKa = 3.7EE150 pKa = 4.53SGVDD154 pKa = 3.16EE155 pKa = 5.08KK156 pKa = 11.73GIFFIKK162 pKa = 10.65YY163 pKa = 8.04GVEE166 pKa = 3.71KK167 pKa = 9.9IYY169 pKa = 10.86YY170 pKa = 10.26LSFADD175 pKa = 3.39EE176 pKa = 4.0ATRR179 pKa = 11.84YY180 pKa = 8.79SRR182 pKa = 11.84KK183 pKa = 10.22GEE185 pKa = 3.96YY186 pKa = 8.74TVHH189 pKa = 6.87FKK191 pKa = 10.0SQRR194 pKa = 11.84HH195 pKa = 5.15SYY197 pKa = 9.41NVSSVSSTSGSPGSPDD213 pKa = 3.05STEE216 pKa = 4.18TNPSSSHH223 pKa = 4.05TRR225 pKa = 11.84GAEE228 pKa = 3.86EE229 pKa = 4.28EE230 pKa = 4.43SPEE233 pKa = 4.01RR234 pKa = 11.84PVRR237 pKa = 11.84SRR239 pKa = 11.84AYY241 pKa = 8.99GQRR244 pKa = 11.84PSTSPRR250 pKa = 11.84VSSRR254 pKa = 11.84RR255 pKa = 11.84GGEE258 pKa = 3.7QGKK261 pKa = 10.16SGTGTDD267 pKa = 3.87SDD269 pKa = 4.32SGLAPPSPGDD279 pKa = 3.35VGSRR283 pKa = 11.84TTQPARR289 pKa = 11.84RR290 pKa = 11.84NQSRR294 pKa = 11.84LRR296 pKa = 11.84VLLQEE301 pKa = 4.48ARR303 pKa = 11.84DD304 pKa = 3.86PLVLCLKK311 pKa = 10.45GGPNQLKK318 pKa = 9.96CLRR321 pKa = 11.84YY322 pKa = 8.85RR323 pKa = 11.84LKK325 pKa = 10.55KK326 pKa = 9.25QHH328 pKa = 6.59HH329 pKa = 6.29KK330 pKa = 10.8LFTKK334 pKa = 10.47ISTTWHH340 pKa = 5.59WVDD343 pKa = 3.09NTSTNRR349 pKa = 11.84VGNARR354 pKa = 11.84MLIQFLTEE362 pKa = 3.57EE363 pKa = 4.24QRR365 pKa = 11.84NHH367 pKa = 6.35FLDD370 pKa = 4.37VIIVPKK376 pKa = 10.31DD377 pKa = 2.89ISVYY381 pKa = 9.83RR382 pKa = 11.84GYY384 pKa = 11.15FKK386 pKa = 11.26GFF388 pKa = 3.26
Molecular weight: 44.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2373 |
92 |
615 |
339.0 |
38.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.436 ± 0.368 | 1.686 ± 0.46 |
6.068 ± 0.426 | 6.321 ± 0.61 |
4.846 ± 0.413 | 5.647 ± 0.585 |
1.938 ± 0.222 | 5.394 ± 0.7 |
4.846 ± 0.799 | 9.524 ± 1.088 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.475 ± 0.223 | 4.888 ± 0.479 |
5.9 ± 0.867 | 5.015 ± 0.64 |
6.068 ± 0.707 | 8.007 ± 1.002 |
6.743 ± 0.283 | 5.942 ± 0.52 |
1.18 ± 0.306 | 3.076 ± 0.324 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
