
Macaca mulatta feces associated virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus macas4
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
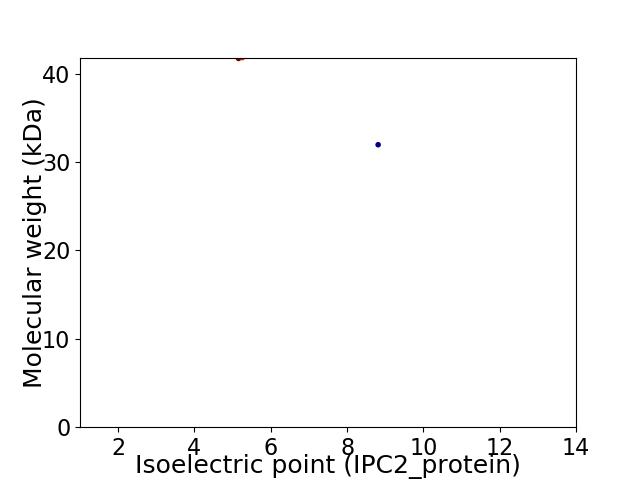
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
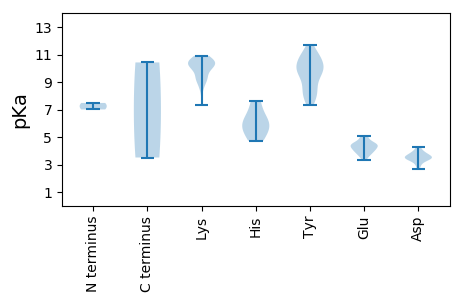
Protein with the lowest isoelectric point:
>tr|A0A1W5PWS6|A0A1W5PWS6_9VIRU Putative capsid protein OS=Macaca mulatta feces associated virus 2 OX=2499224 PE=4 SV=1
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.5TYY9 pKa = 10.76DD10 pKa = 3.38LSTKK14 pKa = 9.04IGKK17 pKa = 8.47MGLVGIHH24 pKa = 5.42TPEE27 pKa = 3.77GRR29 pKa = 11.84LVFKK33 pKa = 10.49QWEE36 pKa = 4.74GFFKK40 pKa = 10.52NYY42 pKa = 9.89RR43 pKa = 11.84KK44 pKa = 9.42IRR46 pKa = 11.84YY47 pKa = 7.99HH48 pKa = 6.01SCDD51 pKa = 2.87VAMACASMLPADD63 pKa = 4.26PLQIGVEE70 pKa = 4.06AGSIAPQDD78 pKa = 3.62MFNPILYY85 pKa = 9.92KK86 pKa = 10.44PVSNDD91 pKa = 3.52SMSTLLNRR99 pKa = 11.84LYY101 pKa = 10.96VGSTAGSPAVAKK113 pKa = 10.87NSVIDD118 pKa = 3.74TNDD121 pKa = 2.94FNFGDD126 pKa = 4.09SQVSQDD132 pKa = 3.43ASGADD137 pKa = 3.52VPVKK141 pKa = 10.67SVGGDD146 pKa = 3.1QFQIYY151 pKa = 10.16YY152 pKa = 10.78GLLSDD157 pKa = 3.49TDD159 pKa = 3.73GWXKK163 pKa = 10.76AMPQSGLEE171 pKa = 4.07MKK173 pKa = 10.48GLYY176 pKa = 9.73PLVFSTLKK184 pKa = 10.18TVGQPSNWSSSTRR197 pKa = 11.84QGIVADD203 pKa = 3.62TAMNLVGEE211 pKa = 4.47SQAYY215 pKa = 9.27QYY217 pKa = 11.05APNVGFIGQYY227 pKa = 8.92MRR229 pKa = 11.84GPAQRR234 pKa = 11.84MPACDD239 pKa = 3.42TYY241 pKa = 11.21LACTGNPNXRR251 pKa = 11.84VEE253 pKa = 4.59GEE255 pKa = 4.34TITIPDD261 pKa = 3.3SRR263 pKa = 11.84XNDD266 pKa = 3.53KK267 pKa = 10.88AYY269 pKa = 7.35PTLSKK274 pKa = 9.95TPTLTRR280 pKa = 11.84VTGPALSNSMAPNLDD295 pKa = 3.52MPDD298 pKa = 3.87CFVGAIVLPPAKK310 pKa = 10.18LNQLYY315 pKa = 10.26YY316 pKa = 10.12RR317 pKa = 11.84LKK319 pKa = 9.08VTWTVEE325 pKa = 4.08FTEE328 pKa = 4.43PRR330 pKa = 11.84PLTDD334 pKa = 3.78LSNWYY339 pKa = 9.71GIQSIGFQSYY349 pKa = 7.87ATDD352 pKa = 3.45YY353 pKa = 9.12ATQTQALQSSGSSVTLGMLDD373 pKa = 3.59TANVDD378 pKa = 3.2AEE380 pKa = 4.48KK381 pKa = 10.91IMDD384 pKa = 3.68GTKK387 pKa = 10.42
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.5TYY9 pKa = 10.76DD10 pKa = 3.38LSTKK14 pKa = 9.04IGKK17 pKa = 8.47MGLVGIHH24 pKa = 5.42TPEE27 pKa = 3.77GRR29 pKa = 11.84LVFKK33 pKa = 10.49QWEE36 pKa = 4.74GFFKK40 pKa = 10.52NYY42 pKa = 9.89RR43 pKa = 11.84KK44 pKa = 9.42IRR46 pKa = 11.84YY47 pKa = 7.99HH48 pKa = 6.01SCDD51 pKa = 2.87VAMACASMLPADD63 pKa = 4.26PLQIGVEE70 pKa = 4.06AGSIAPQDD78 pKa = 3.62MFNPILYY85 pKa = 9.92KK86 pKa = 10.44PVSNDD91 pKa = 3.52SMSTLLNRR99 pKa = 11.84LYY101 pKa = 10.96VGSTAGSPAVAKK113 pKa = 10.87NSVIDD118 pKa = 3.74TNDD121 pKa = 2.94FNFGDD126 pKa = 4.09SQVSQDD132 pKa = 3.43ASGADD137 pKa = 3.52VPVKK141 pKa = 10.67SVGGDD146 pKa = 3.1QFQIYY151 pKa = 10.16YY152 pKa = 10.78GLLSDD157 pKa = 3.49TDD159 pKa = 3.73GWXKK163 pKa = 10.76AMPQSGLEE171 pKa = 4.07MKK173 pKa = 10.48GLYY176 pKa = 9.73PLVFSTLKK184 pKa = 10.18TVGQPSNWSSSTRR197 pKa = 11.84QGIVADD203 pKa = 3.62TAMNLVGEE211 pKa = 4.47SQAYY215 pKa = 9.27QYY217 pKa = 11.05APNVGFIGQYY227 pKa = 8.92MRR229 pKa = 11.84GPAQRR234 pKa = 11.84MPACDD239 pKa = 3.42TYY241 pKa = 11.21LACTGNPNXRR251 pKa = 11.84VEE253 pKa = 4.59GEE255 pKa = 4.34TITIPDD261 pKa = 3.3SRR263 pKa = 11.84XNDD266 pKa = 3.53KK267 pKa = 10.88AYY269 pKa = 7.35PTLSKK274 pKa = 9.95TPTLTRR280 pKa = 11.84VTGPALSNSMAPNLDD295 pKa = 3.52MPDD298 pKa = 3.87CFVGAIVLPPAKK310 pKa = 10.18LNQLYY315 pKa = 10.26YY316 pKa = 10.12RR317 pKa = 11.84LKK319 pKa = 9.08VTWTVEE325 pKa = 4.08FTEE328 pKa = 4.43PRR330 pKa = 11.84PLTDD334 pKa = 3.78LSNWYY339 pKa = 9.71GIQSIGFQSYY349 pKa = 7.87ATDD352 pKa = 3.45YY353 pKa = 9.12ATQTQALQSSGSSVTLGMLDD373 pKa = 3.59TANVDD378 pKa = 3.2AEE380 pKa = 4.48KK381 pKa = 10.91IMDD384 pKa = 3.68GTKK387 pKa = 10.42
Molecular weight: 41.73 kDa
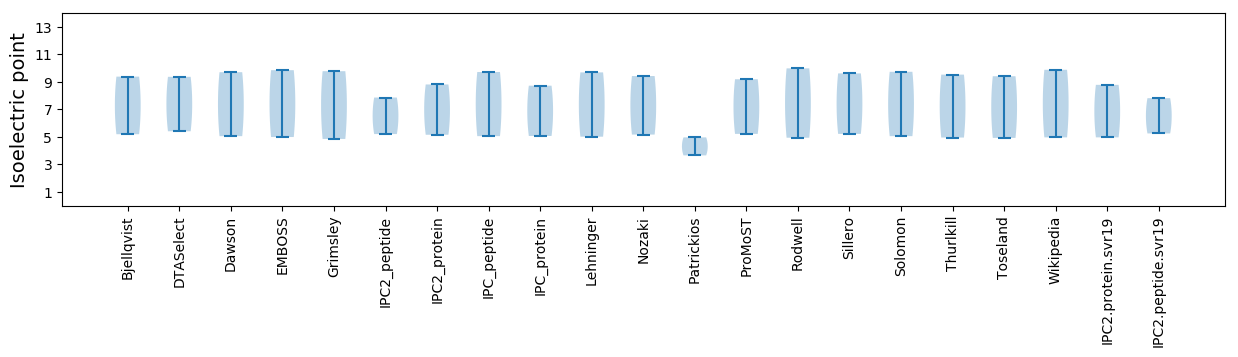
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5PWS6|A0A1W5PWS6_9VIRU Putative capsid protein OS=Macaca mulatta feces associated virus 2 OX=2499224 PE=4 SV=1
MM1 pKa = 7.48TKK3 pKa = 9.52TVMITMEE10 pKa = 3.55RR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 4.31TYY15 pKa = 10.48KK16 pKa = 10.2EE17 pKa = 4.0WNWFLFAIRR26 pKa = 11.84KK27 pKa = 9.31LDD29 pKa = 3.21IHH31 pKa = 6.5KK32 pKa = 10.0WIIAAEE38 pKa = 4.05EE39 pKa = 4.3GKK41 pKa = 10.63GGYY44 pKa = 8.49KK45 pKa = 9.74HH46 pKa = 4.74WQIRR50 pKa = 11.84IASRR54 pKa = 11.84FDD56 pKa = 3.49AKK58 pKa = 9.14WWTRR62 pKa = 11.84ALLIQFGQRR71 pKa = 11.84SIHH74 pKa = 5.82TEE76 pKa = 3.63NASNTWTYY84 pKa = 8.55EE85 pKa = 3.9AKK87 pKa = 10.2EE88 pKa = 4.24GCYY91 pKa = 9.56LASWDD96 pKa = 3.92TLEE99 pKa = 5.01VRR101 pKa = 11.84QQRR104 pKa = 11.84FGKK107 pKa = 9.39LRR109 pKa = 11.84KK110 pKa = 7.32VQKK113 pKa = 10.05YY114 pKa = 9.74ALNVLEE120 pKa = 4.42GTNDD124 pKa = 3.39RR125 pKa = 11.84QVMVWVDD132 pKa = 3.31EE133 pKa = 4.44EE134 pKa = 4.76GNSGKK139 pKa = 10.11SWLIGHH145 pKa = 7.63LYY147 pKa = 8.26EE148 pKa = 5.09TGQAYY153 pKa = 7.68YY154 pKa = 10.48APPYY158 pKa = 8.19LTSVKK163 pKa = 10.38EE164 pKa = 4.12IIQTIASLAVKK175 pKa = 10.36DD176 pKa = 4.29RR177 pKa = 11.84DD178 pKa = 4.0AGNPPKK184 pKa = 10.32KK185 pKa = 10.04YY186 pKa = 10.86ALIDD190 pKa = 3.53IPRR193 pKa = 11.84SWKK196 pKa = 9.01WSNEE200 pKa = 3.36LYY202 pKa = 10.17TAIEE206 pKa = 4.52AIKK209 pKa = 10.37DD210 pKa = 3.7GLIMEE215 pKa = 5.1PRR217 pKa = 11.84YY218 pKa = 9.79SAQPINIKK226 pKa = 9.97GIKK229 pKa = 9.32VLVVTNTRR237 pKa = 11.84PKK239 pKa = 10.45LDD241 pKa = 3.93KK242 pKa = 10.63LSKK245 pKa = 10.47DD246 pKa = 2.67RR247 pKa = 11.84WEE249 pKa = 3.85IFEE252 pKa = 4.4WDD254 pKa = 3.79NNPTIDD260 pKa = 3.32TYY262 pKa = 11.68LRR264 pKa = 11.84GAQEE268 pKa = 3.74KK269 pKa = 8.56RR270 pKa = 11.84TLL272 pKa = 3.51
MM1 pKa = 7.48TKK3 pKa = 9.52TVMITMEE10 pKa = 3.55RR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 4.31TYY15 pKa = 10.48KK16 pKa = 10.2EE17 pKa = 4.0WNWFLFAIRR26 pKa = 11.84KK27 pKa = 9.31LDD29 pKa = 3.21IHH31 pKa = 6.5KK32 pKa = 10.0WIIAAEE38 pKa = 4.05EE39 pKa = 4.3GKK41 pKa = 10.63GGYY44 pKa = 8.49KK45 pKa = 9.74HH46 pKa = 4.74WQIRR50 pKa = 11.84IASRR54 pKa = 11.84FDD56 pKa = 3.49AKK58 pKa = 9.14WWTRR62 pKa = 11.84ALLIQFGQRR71 pKa = 11.84SIHH74 pKa = 5.82TEE76 pKa = 3.63NASNTWTYY84 pKa = 8.55EE85 pKa = 3.9AKK87 pKa = 10.2EE88 pKa = 4.24GCYY91 pKa = 9.56LASWDD96 pKa = 3.92TLEE99 pKa = 5.01VRR101 pKa = 11.84QQRR104 pKa = 11.84FGKK107 pKa = 9.39LRR109 pKa = 11.84KK110 pKa = 7.32VQKK113 pKa = 10.05YY114 pKa = 9.74ALNVLEE120 pKa = 4.42GTNDD124 pKa = 3.39RR125 pKa = 11.84QVMVWVDD132 pKa = 3.31EE133 pKa = 4.44EE134 pKa = 4.76GNSGKK139 pKa = 10.11SWLIGHH145 pKa = 7.63LYY147 pKa = 8.26EE148 pKa = 5.09TGQAYY153 pKa = 7.68YY154 pKa = 10.48APPYY158 pKa = 8.19LTSVKK163 pKa = 10.38EE164 pKa = 4.12IIQTIASLAVKK175 pKa = 10.36DD176 pKa = 4.29RR177 pKa = 11.84DD178 pKa = 4.0AGNPPKK184 pKa = 10.32KK185 pKa = 10.04YY186 pKa = 10.86ALIDD190 pKa = 3.53IPRR193 pKa = 11.84SWKK196 pKa = 9.01WSNEE200 pKa = 3.36LYY202 pKa = 10.17TAIEE206 pKa = 4.52AIKK209 pKa = 10.37DD210 pKa = 3.7GLIMEE215 pKa = 5.1PRR217 pKa = 11.84YY218 pKa = 9.79SAQPINIKK226 pKa = 9.97GIKK229 pKa = 9.32VLVVTNTRR237 pKa = 11.84PKK239 pKa = 10.45LDD241 pKa = 3.93KK242 pKa = 10.63LSKK245 pKa = 10.47DD246 pKa = 2.67RR247 pKa = 11.84WEE249 pKa = 3.85IFEE252 pKa = 4.4WDD254 pKa = 3.79NNPTIDD260 pKa = 3.32TYY262 pKa = 11.68LRR264 pKa = 11.84GAQEE268 pKa = 3.74KK269 pKa = 8.56RR270 pKa = 11.84TLL272 pKa = 3.51
Molecular weight: 31.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
659 |
272 |
387 |
329.5 |
36.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.436 ± 0.045 | 0.91 ± 0.296 |
5.615 ± 0.455 | 4.704 ± 1.443 |
2.883 ± 0.369 | 7.132 ± 0.881 |
0.91 ± 0.305 | 5.918 ± 1.383 |
6.373 ± 1.335 | 7.739 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.035 ± 0.652 | 4.552 ± 0.077 |
5.159 ± 1.008 | 4.704 ± 0.36 |
4.552 ± 1.125 | 6.98 ± 1.4 |
7.436 ± 0.245 | 5.918 ± 0.821 |
2.883 ± 1.234 | 4.704 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
