
Guangxi orbivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; unclassified Orbivirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
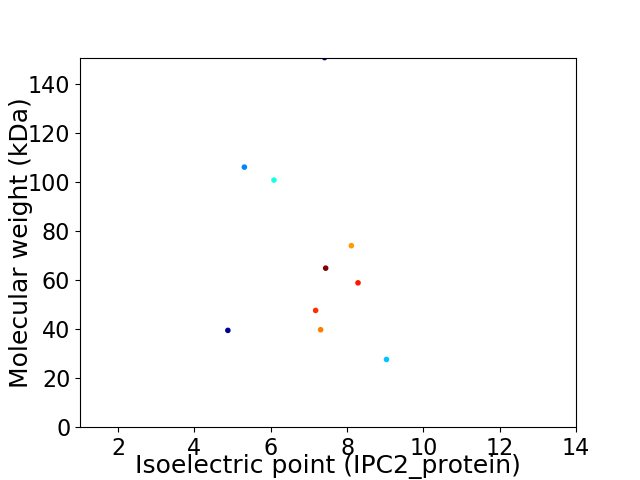
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346RV23|A0A346RV23_9REOV NTPase OS=Guangxi orbivirus OX=2306813 GN=VP6 PE=4 SV=1
MM1 pKa = 7.34EE2 pKa = 6.05GIYY5 pKa = 10.76ARR7 pKa = 11.84TFCYY11 pKa = 9.82MEE13 pKa = 4.34CLGSNRR19 pKa = 11.84DD20 pKa = 3.13PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 9.97IPSDD31 pKa = 3.57GFPLFVTRR39 pKa = 11.84FNSTTDD45 pKa = 3.25RR46 pKa = 11.84PITQTPSTIEE56 pKa = 3.55EE57 pKa = 4.22HH58 pKa = 6.83RR59 pKa = 11.84NCFYY63 pKa = 11.35AGLDD67 pKa = 3.69LLIAALGVNFNYY79 pKa = 10.45NLPDD83 pKa = 3.43YY84 pKa = 9.18TPNVQVLSLLARR96 pKa = 11.84EE97 pKa = 4.27DD98 pKa = 3.93LPYY101 pKa = 9.88STHH104 pKa = 7.13AYY106 pKa = 9.12IRR108 pKa = 11.84AQRR111 pKa = 11.84IVNEE115 pKa = 3.99PAAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FYY126 pKa = 10.91TPWPDD131 pKa = 3.15VNFIYY136 pKa = 10.35PPATPYY142 pKa = 9.43RR143 pKa = 11.84TGADD147 pKa = 3.96EE148 pKa = 4.45PQPMCTGPQSMEE160 pKa = 3.62VTLNANEE167 pKa = 4.47DD168 pKa = 3.86LEE170 pKa = 4.45ITNTVMPNYY179 pKa = 9.17PDD181 pKa = 4.51VVDD184 pKa = 4.6TCITWYY190 pKa = 9.29TLSHH194 pKa = 6.02FRR196 pKa = 11.84GAGGALIEE204 pKa = 5.03GSQLCNLMIGEE215 pKa = 4.93DD216 pKa = 3.73EE217 pKa = 4.5VEE219 pKa = 4.24AGTEE223 pKa = 4.01IVVSADD229 pKa = 3.04AQIRR233 pKa = 11.84VANQSAVNAGMVHH246 pKa = 6.94IDD248 pKa = 3.33VRR250 pKa = 11.84WFTRR254 pKa = 11.84AQPSEE259 pKa = 4.81DD260 pKa = 3.4IYY262 pKa = 11.35DD263 pKa = 3.75TMRR266 pKa = 11.84VDD268 pKa = 3.18IASSYY273 pKa = 10.9SFHH276 pKa = 6.08SQQWYY281 pKa = 9.91DD282 pKa = 3.18LRR284 pKa = 11.84TYY286 pKa = 10.76LLRR289 pKa = 11.84VMGLPAHH296 pKa = 6.45MPTTEE301 pKa = 4.97PIRR304 pKa = 11.84NPQRR308 pKa = 11.84ILTLALISRR317 pKa = 11.84LLDD320 pKa = 3.02IYY322 pKa = 10.56VAHH325 pKa = 6.74SPIDD329 pKa = 3.41LTAIQGANGQAGIPDD344 pKa = 3.95AALDD348 pKa = 3.57ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.7
MM1 pKa = 7.34EE2 pKa = 6.05GIYY5 pKa = 10.76ARR7 pKa = 11.84TFCYY11 pKa = 9.82MEE13 pKa = 4.34CLGSNRR19 pKa = 11.84DD20 pKa = 3.13PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 9.97IPSDD31 pKa = 3.57GFPLFVTRR39 pKa = 11.84FNSTTDD45 pKa = 3.25RR46 pKa = 11.84PITQTPSTIEE56 pKa = 3.55EE57 pKa = 4.22HH58 pKa = 6.83RR59 pKa = 11.84NCFYY63 pKa = 11.35AGLDD67 pKa = 3.69LLIAALGVNFNYY79 pKa = 10.45NLPDD83 pKa = 3.43YY84 pKa = 9.18TPNVQVLSLLARR96 pKa = 11.84EE97 pKa = 4.27DD98 pKa = 3.93LPYY101 pKa = 9.88STHH104 pKa = 7.13AYY106 pKa = 9.12IRR108 pKa = 11.84AQRR111 pKa = 11.84IVNEE115 pKa = 3.99PAAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FYY126 pKa = 10.91TPWPDD131 pKa = 3.15VNFIYY136 pKa = 10.35PPATPYY142 pKa = 9.43RR143 pKa = 11.84TGADD147 pKa = 3.96EE148 pKa = 4.45PQPMCTGPQSMEE160 pKa = 3.62VTLNANEE167 pKa = 4.47DD168 pKa = 3.86LEE170 pKa = 4.45ITNTVMPNYY179 pKa = 9.17PDD181 pKa = 4.51VVDD184 pKa = 4.6TCITWYY190 pKa = 9.29TLSHH194 pKa = 6.02FRR196 pKa = 11.84GAGGALIEE204 pKa = 5.03GSQLCNLMIGEE215 pKa = 4.93DD216 pKa = 3.73EE217 pKa = 4.5VEE219 pKa = 4.24AGTEE223 pKa = 4.01IVVSADD229 pKa = 3.04AQIRR233 pKa = 11.84VANQSAVNAGMVHH246 pKa = 6.94IDD248 pKa = 3.33VRR250 pKa = 11.84WFTRR254 pKa = 11.84AQPSEE259 pKa = 4.81DD260 pKa = 3.4IYY262 pKa = 11.35DD263 pKa = 3.75TMRR266 pKa = 11.84VDD268 pKa = 3.18IASSYY273 pKa = 10.9SFHH276 pKa = 6.08SQQWYY281 pKa = 9.91DD282 pKa = 3.18LRR284 pKa = 11.84TYY286 pKa = 10.76LLRR289 pKa = 11.84VMGLPAHH296 pKa = 6.45MPTTEE301 pKa = 4.97PIRR304 pKa = 11.84NPQRR308 pKa = 11.84ILTLALISRR317 pKa = 11.84LLDD320 pKa = 3.02IYY322 pKa = 10.56VAHH325 pKa = 6.74SPIDD329 pKa = 3.41LTAIQGANGQAGIPDD344 pKa = 3.95AALDD348 pKa = 3.57ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.7
Molecular weight: 39.52 kDa
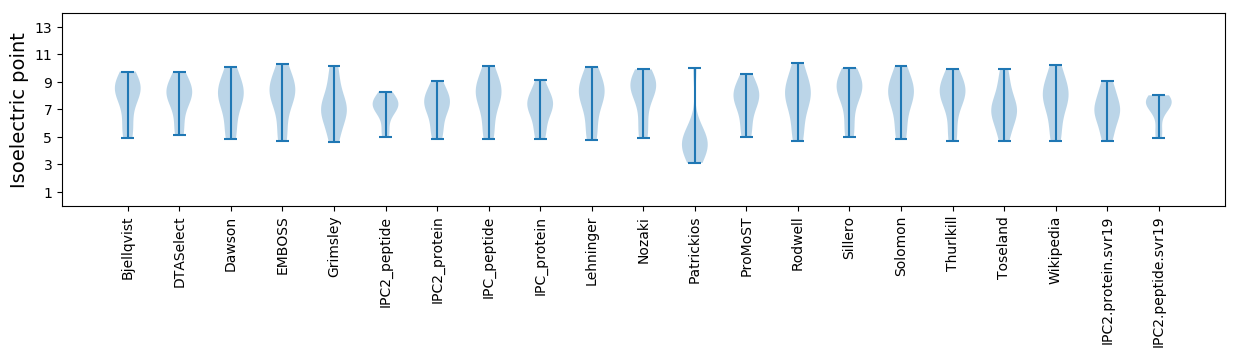
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346RV24|A0A346RV24_9REOV Non-structural protein NS3 OS=Guangxi orbivirus OX=2306813 GN=NS3 PE=3 SV=1
MM1 pKa = 7.27HH2 pKa = 7.44AAISSAKK9 pKa = 10.04RR10 pKa = 11.84EE11 pKa = 4.02NDD13 pKa = 3.18PTSLVVVPATAPRR26 pKa = 11.84LEE28 pKa = 4.38EE29 pKa = 4.1VGEE32 pKa = 4.24VPLLDD37 pKa = 3.32KK38 pKa = 10.59RR39 pKa = 11.84VRR41 pKa = 11.84FALPDD46 pKa = 3.77VNVSTAKK53 pKa = 10.38SVALDD58 pKa = 3.41VLANALGGGTGTDD71 pKa = 3.15EE72 pKa = 4.02ITRR75 pKa = 11.84QEE77 pKa = 3.78RR78 pKa = 11.84SAYY81 pKa = 9.34GAAAQALNEE90 pKa = 4.19DD91 pKa = 4.35ANTKK95 pKa = 7.86VLRR98 pKa = 11.84VYY100 pKa = 10.34MNAQIMPRR108 pKa = 11.84LKK110 pKa = 10.84GKK112 pKa = 10.07LRR114 pKa = 11.84TANVKK119 pKa = 9.8YY120 pKa = 10.31RR121 pKa = 11.84FWLALEE127 pKa = 4.03ITLALISTAIFSIMAVTEE145 pKa = 3.95AEE147 pKa = 4.1KK148 pKa = 10.01TVNSTIEE155 pKa = 3.92RR156 pKa = 11.84VTGLKK161 pKa = 10.37VGLSAFSLICTGALLFASRR180 pKa = 11.84HH181 pKa = 5.18AGSLNEE187 pKa = 3.57FRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84LRR193 pKa = 11.84RR194 pKa = 11.84EE195 pKa = 3.66VVKK198 pKa = 10.81RR199 pKa = 11.84EE200 pKa = 3.97TYY202 pKa = 10.65NFVANNHH209 pKa = 6.15EE210 pKa = 4.27IGRR213 pKa = 11.84LQIGTEE219 pKa = 3.99TRR221 pKa = 11.84GDD223 pKa = 3.48AGQSRR228 pKa = 11.84MNVVGNPAWISAPGNAAHH246 pKa = 7.35AEE248 pKa = 4.3GWQLSFVV255 pKa = 3.83
MM1 pKa = 7.27HH2 pKa = 7.44AAISSAKK9 pKa = 10.04RR10 pKa = 11.84EE11 pKa = 4.02NDD13 pKa = 3.18PTSLVVVPATAPRR26 pKa = 11.84LEE28 pKa = 4.38EE29 pKa = 4.1VGEE32 pKa = 4.24VPLLDD37 pKa = 3.32KK38 pKa = 10.59RR39 pKa = 11.84VRR41 pKa = 11.84FALPDD46 pKa = 3.77VNVSTAKK53 pKa = 10.38SVALDD58 pKa = 3.41VLANALGGGTGTDD71 pKa = 3.15EE72 pKa = 4.02ITRR75 pKa = 11.84QEE77 pKa = 3.78RR78 pKa = 11.84SAYY81 pKa = 9.34GAAAQALNEE90 pKa = 4.19DD91 pKa = 4.35ANTKK95 pKa = 7.86VLRR98 pKa = 11.84VYY100 pKa = 10.34MNAQIMPRR108 pKa = 11.84LKK110 pKa = 10.84GKK112 pKa = 10.07LRR114 pKa = 11.84TANVKK119 pKa = 9.8YY120 pKa = 10.31RR121 pKa = 11.84FWLALEE127 pKa = 4.03ITLALISTAIFSIMAVTEE145 pKa = 3.95AEE147 pKa = 4.1KK148 pKa = 10.01TVNSTIEE155 pKa = 3.92RR156 pKa = 11.84VTGLKK161 pKa = 10.37VGLSAFSLICTGALLFASRR180 pKa = 11.84HH181 pKa = 5.18AGSLNEE187 pKa = 3.57FRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84LRR193 pKa = 11.84RR194 pKa = 11.84EE195 pKa = 3.66VVKK198 pKa = 10.81RR199 pKa = 11.84EE200 pKa = 3.97TYY202 pKa = 10.65NFVANNHH209 pKa = 6.15EE210 pKa = 4.27IGRR213 pKa = 11.84LQIGTEE219 pKa = 3.99TRR221 pKa = 11.84GDD223 pKa = 3.48AGQSRR228 pKa = 11.84MNVVGNPAWISAPGNAAHH246 pKa = 7.35AEE248 pKa = 4.3GWQLSFVV255 pKa = 3.83
Molecular weight: 27.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6265 |
255 |
1316 |
626.5 |
71.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.496 ± 0.553 | 1.468 ± 0.258 |
5.443 ± 0.377 | 6.864 ± 0.408 |
3.847 ± 0.278 | 5.379 ± 0.297 |
2.203 ± 0.293 | 6.736 ± 0.357 |
5.698 ± 0.541 | 9.21 ± 0.315 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.384 ± 0.207 | 4.964 ± 0.286 |
3.911 ± 0.386 | 3.272 ± 0.193 |
6.672 ± 0.202 | 7.39 ± 0.427 |
5.666 ± 0.331 | 6.991 ± 0.578 |
1.038 ± 0.218 | 3.368 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
