
Klebsiella phage 1611E-K2-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Jedunavirus; unclassified Jedunavirus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
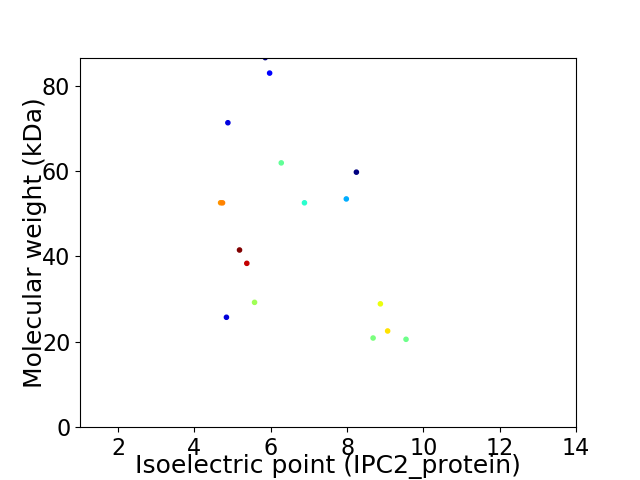
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A3S6PUF3|A0A3S6PUF3_9CAUD Phage_Mu_F domain-containing protein OS=Klebsiella phage 1611E-K2-1 OX=2047786 GN=1611EK21_7 PE=4 SV=1
MM1 pKa = 7.49TIIKK5 pKa = 10.09RR6 pKa = 11.84ADD8 pKa = 3.26LGRR11 pKa = 11.84PLTWDD16 pKa = 3.42EE17 pKa = 4.98LDD19 pKa = 5.79DD20 pKa = 5.05NFQQVDD26 pKa = 4.31DD27 pKa = 4.24LTAAASAAVLSASASATAAAGSATNSLNSANSAASSADD65 pKa = 3.35DD66 pKa = 3.63AAASATVAINALMNSTFEE84 pKa = 4.08PADD87 pKa = 3.51FDD89 pKa = 4.06FTSGGTLDD97 pKa = 3.38STDD100 pKa = 3.43RR101 pKa = 11.84NKK103 pKa = 10.89AVYY106 pKa = 10.32NPADD110 pKa = 3.86NNWYY114 pKa = 8.76SWSGILPKK122 pKa = 10.14IVTAATDD129 pKa = 3.6PTADD133 pKa = 4.05SNWKK137 pKa = 9.38PRR139 pKa = 11.84TDD141 pKa = 3.14QLLRR145 pKa = 11.84QNLASSVIPGTSLVTHH161 pKa = 6.92SDD163 pKa = 3.98GIHH166 pKa = 6.46LDD168 pKa = 3.6DD169 pKa = 5.37YY170 pKa = 11.7IEE172 pKa = 4.17IFNRR176 pKa = 11.84RR177 pKa = 11.84TKK179 pKa = 10.78FIMPEE184 pKa = 4.1DD185 pKa = 3.96FPGTDD190 pKa = 3.35TEE192 pKa = 4.38QLQSALSYY200 pKa = 10.84AKK202 pKa = 10.08SNRR205 pKa = 11.84VNVVLQAGKK214 pKa = 8.25TYY216 pKa = 10.81YY217 pKa = 9.2VTGSQGLEE225 pKa = 3.68VDD227 pKa = 3.66LGYY230 pKa = 11.4YY231 pKa = 10.34SFEE234 pKa = 4.27SPNGIAYY241 pKa = 9.24IDD243 pKa = 3.84FTGCTATYY251 pKa = 9.12CLWVHH256 pKa = 6.33SSRR259 pKa = 11.84PYY261 pKa = 10.05PDD263 pKa = 3.78GSEE266 pKa = 3.99NHH268 pKa = 5.81CTSMRR273 pKa = 11.84GIKK276 pKa = 9.86FKK278 pKa = 11.27SSVKK282 pKa = 10.76GIGQRR287 pKa = 11.84LLLTGNNNDD296 pKa = 4.15SSNGTYY302 pKa = 10.71NGDD305 pKa = 3.76CKK307 pKa = 10.58IEE309 pKa = 3.79NCMFSTADD317 pKa = 3.22IVLGASNSTWRR328 pKa = 11.84YY329 pKa = 9.87KK330 pKa = 10.6FINCGFMMEE339 pKa = 4.62STGGTYY345 pKa = 11.09AMHH348 pKa = 6.71FPAGISDD355 pKa = 3.7SGEE358 pKa = 3.71SVTFQNCKK366 pKa = 9.49IFDD369 pKa = 3.78MKK371 pKa = 10.86GCPILVEE378 pKa = 4.28CASFAIGMPGTSVLNTPIKK397 pKa = 9.55ITGSGAMVIMDD408 pKa = 4.09SAANIEE414 pKa = 4.36NPGASAWYY422 pKa = 9.56RR423 pKa = 11.84YY424 pKa = 10.67GEE426 pKa = 4.23VTGTGARR433 pKa = 11.84LILNGCTLVCNNPSLQTKK451 pKa = 8.88PLFYY455 pKa = 10.85VGANAFIDD463 pKa = 3.75VTLVKK468 pKa = 10.15TPGNDD473 pKa = 3.2YY474 pKa = 10.97LFQNGDD480 pKa = 2.96EE481 pKa = 4.31GLRR484 pKa = 11.84TFVEE488 pKa = 4.03GDD490 pKa = 3.68GYY492 pKa = 9.91VTASHH497 pKa = 7.33CIGDD501 pKa = 3.77ILSGVGNIPLHH512 pKa = 6.43KK513 pKa = 10.46SLNPTLNPGFEE524 pKa = 4.31TGDD527 pKa = 3.41LSSWSFNNQGSASQTCVVGTAYY549 pKa = 10.0KK550 pKa = 9.32KK551 pKa = 8.9TGTYY555 pKa = 8.69GARR558 pKa = 11.84MTSFGSLSCFLTQKK572 pKa = 10.58VKK574 pKa = 9.22VTQHH578 pKa = 5.65GYY580 pKa = 11.18YY581 pKa = 9.24STTCQINTITAGTGTTAGSLTITFYY606 pKa = 11.35NRR608 pKa = 11.84DD609 pKa = 3.51GNALQAGASSNFTNTPSGWQSVGRR633 pKa = 11.84FIQGRR638 pKa = 11.84VPQAAEE644 pKa = 3.78YY645 pKa = 10.57CEE647 pKa = 4.16VSFRR651 pKa = 11.84CRR653 pKa = 11.84EE654 pKa = 3.89GAVIDD659 pKa = 3.67VDD661 pKa = 3.64NFIINFTT668 pKa = 3.83
MM1 pKa = 7.49TIIKK5 pKa = 10.09RR6 pKa = 11.84ADD8 pKa = 3.26LGRR11 pKa = 11.84PLTWDD16 pKa = 3.42EE17 pKa = 4.98LDD19 pKa = 5.79DD20 pKa = 5.05NFQQVDD26 pKa = 4.31DD27 pKa = 4.24LTAAASAAVLSASASATAAAGSATNSLNSANSAASSADD65 pKa = 3.35DD66 pKa = 3.63AAASATVAINALMNSTFEE84 pKa = 4.08PADD87 pKa = 3.51FDD89 pKa = 4.06FTSGGTLDD97 pKa = 3.38STDD100 pKa = 3.43RR101 pKa = 11.84NKK103 pKa = 10.89AVYY106 pKa = 10.32NPADD110 pKa = 3.86NNWYY114 pKa = 8.76SWSGILPKK122 pKa = 10.14IVTAATDD129 pKa = 3.6PTADD133 pKa = 4.05SNWKK137 pKa = 9.38PRR139 pKa = 11.84TDD141 pKa = 3.14QLLRR145 pKa = 11.84QNLASSVIPGTSLVTHH161 pKa = 6.92SDD163 pKa = 3.98GIHH166 pKa = 6.46LDD168 pKa = 3.6DD169 pKa = 5.37YY170 pKa = 11.7IEE172 pKa = 4.17IFNRR176 pKa = 11.84RR177 pKa = 11.84TKK179 pKa = 10.78FIMPEE184 pKa = 4.1DD185 pKa = 3.96FPGTDD190 pKa = 3.35TEE192 pKa = 4.38QLQSALSYY200 pKa = 10.84AKK202 pKa = 10.08SNRR205 pKa = 11.84VNVVLQAGKK214 pKa = 8.25TYY216 pKa = 10.81YY217 pKa = 9.2VTGSQGLEE225 pKa = 3.68VDD227 pKa = 3.66LGYY230 pKa = 11.4YY231 pKa = 10.34SFEE234 pKa = 4.27SPNGIAYY241 pKa = 9.24IDD243 pKa = 3.84FTGCTATYY251 pKa = 9.12CLWVHH256 pKa = 6.33SSRR259 pKa = 11.84PYY261 pKa = 10.05PDD263 pKa = 3.78GSEE266 pKa = 3.99NHH268 pKa = 5.81CTSMRR273 pKa = 11.84GIKK276 pKa = 9.86FKK278 pKa = 11.27SSVKK282 pKa = 10.76GIGQRR287 pKa = 11.84LLLTGNNNDD296 pKa = 4.15SSNGTYY302 pKa = 10.71NGDD305 pKa = 3.76CKK307 pKa = 10.58IEE309 pKa = 3.79NCMFSTADD317 pKa = 3.22IVLGASNSTWRR328 pKa = 11.84YY329 pKa = 9.87KK330 pKa = 10.6FINCGFMMEE339 pKa = 4.62STGGTYY345 pKa = 11.09AMHH348 pKa = 6.71FPAGISDD355 pKa = 3.7SGEE358 pKa = 3.71SVTFQNCKK366 pKa = 9.49IFDD369 pKa = 3.78MKK371 pKa = 10.86GCPILVEE378 pKa = 4.28CASFAIGMPGTSVLNTPIKK397 pKa = 9.55ITGSGAMVIMDD408 pKa = 4.09SAANIEE414 pKa = 4.36NPGASAWYY422 pKa = 9.56RR423 pKa = 11.84YY424 pKa = 10.67GEE426 pKa = 4.23VTGTGARR433 pKa = 11.84LILNGCTLVCNNPSLQTKK451 pKa = 8.88PLFYY455 pKa = 10.85VGANAFIDD463 pKa = 3.75VTLVKK468 pKa = 10.15TPGNDD473 pKa = 3.2YY474 pKa = 10.97LFQNGDD480 pKa = 2.96EE481 pKa = 4.31GLRR484 pKa = 11.84TFVEE488 pKa = 4.03GDD490 pKa = 3.68GYY492 pKa = 9.91VTASHH497 pKa = 7.33CIGDD501 pKa = 3.77ILSGVGNIPLHH512 pKa = 6.43KK513 pKa = 10.46SLNPTLNPGFEE524 pKa = 4.31TGDD527 pKa = 3.41LSSWSFNNQGSASQTCVVGTAYY549 pKa = 10.0KK550 pKa = 9.32KK551 pKa = 8.9TGTYY555 pKa = 8.69GARR558 pKa = 11.84MTSFGSLSCFLTQKK572 pKa = 10.58VKK574 pKa = 9.22VTQHH578 pKa = 5.65GYY580 pKa = 11.18YY581 pKa = 9.24STTCQINTITAGTGTTAGSLTITFYY606 pKa = 11.35NRR608 pKa = 11.84DD609 pKa = 3.51GNALQAGASSNFTNTPSGWQSVGRR633 pKa = 11.84FIQGRR638 pKa = 11.84VPQAAEE644 pKa = 3.78YY645 pKa = 10.57CEE647 pKa = 4.16VSFRR651 pKa = 11.84CRR653 pKa = 11.84EE654 pKa = 3.89GAVIDD659 pKa = 3.67VDD661 pKa = 3.64NFIINFTT668 pKa = 3.83
Molecular weight: 71.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S6PTV9|A0A3S6PTV9_9CAUD Putative DNA polymerase OS=Klebsiella phage 1611E-K2-1 OX=2047786 GN=1611EK21_13 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84AFRR5 pKa = 11.84FPRR8 pKa = 11.84SEE10 pKa = 3.74ARR12 pKa = 11.84HH13 pKa = 6.46DD14 pKa = 3.44VDD16 pKa = 4.37QPVGGVKK23 pKa = 10.06VCFSSLSHH31 pKa = 6.35VYY33 pKa = 10.08KK34 pKa = 10.63ISDD37 pKa = 3.36AVRR40 pKa = 11.84RR41 pKa = 11.84ASNPRR46 pKa = 11.84LLAMINEE53 pKa = 4.39SAMFGDD59 pKa = 3.8MMSRR63 pKa = 11.84FAKK66 pKa = 10.49SFLLSTLTLPLLSKK80 pKa = 10.83SRR82 pKa = 11.84IGVDD86 pKa = 3.22NSSMSLSSSFASISLDD102 pKa = 3.84KK103 pKa = 11.14LIIWSTGNSRR113 pKa = 11.84PFVTARR119 pKa = 11.84YY120 pKa = 8.8VLRR123 pKa = 11.84NRR125 pKa = 11.84SAVSHH130 pKa = 6.02HH131 pKa = 5.88VCAFRR136 pKa = 11.84LAKK139 pKa = 10.21KK140 pKa = 10.25SSLLMRR146 pKa = 11.84VSPYY150 pKa = 10.55FFFGSDD156 pKa = 3.52TLPPALNLKK165 pKa = 8.81CLCHH169 pKa = 6.3PAAFRR174 pKa = 11.84CEE176 pKa = 4.25PVPAPTPILSS186 pKa = 3.38
MM1 pKa = 7.62RR2 pKa = 11.84AFRR5 pKa = 11.84FPRR8 pKa = 11.84SEE10 pKa = 3.74ARR12 pKa = 11.84HH13 pKa = 6.46DD14 pKa = 3.44VDD16 pKa = 4.37QPVGGVKK23 pKa = 10.06VCFSSLSHH31 pKa = 6.35VYY33 pKa = 10.08KK34 pKa = 10.63ISDD37 pKa = 3.36AVRR40 pKa = 11.84RR41 pKa = 11.84ASNPRR46 pKa = 11.84LLAMINEE53 pKa = 4.39SAMFGDD59 pKa = 3.8MMSRR63 pKa = 11.84FAKK66 pKa = 10.49SFLLSTLTLPLLSKK80 pKa = 10.83SRR82 pKa = 11.84IGVDD86 pKa = 3.22NSSMSLSSSFASISLDD102 pKa = 3.84KK103 pKa = 11.14LIIWSTGNSRR113 pKa = 11.84PFVTARR119 pKa = 11.84YY120 pKa = 8.8VLRR123 pKa = 11.84NRR125 pKa = 11.84SAVSHH130 pKa = 6.02HH131 pKa = 5.88VCAFRR136 pKa = 11.84LAKK139 pKa = 10.21KK140 pKa = 10.25SSLLMRR146 pKa = 11.84VSPYY150 pKa = 10.55FFFGSDD156 pKa = 3.52TLPPALNLKK165 pKa = 8.81CLCHH169 pKa = 6.3PAAFRR174 pKa = 11.84CEE176 pKa = 4.25PVPAPTPILSS186 pKa = 3.38
Molecular weight: 20.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7239 |
186 |
785 |
425.8 |
47.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.551 ± 0.665 | 1.63 ± 0.246 |
6.382 ± 0.337 | 4.835 ± 0.399 |
3.896 ± 0.286 | 7.888 ± 0.62 |
2.017 ± 0.311 | 5.498 ± 0.21 |
5.318 ± 0.436 | 6.879 ± 0.295 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.79 ± 0.244 | 5.415 ± 0.493 |
4.006 ± 0.225 | 4.103 ± 0.428 |
5.484 ± 0.35 | 6.727 ± 0.522 |
6.824 ± 0.425 | 6.797 ± 0.329 |
1.533 ± 0.123 | 3.426 ± 0.225 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
