
Streptomyces phage ToastyFinz
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Raleighvirus; unclassified Raleighvirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
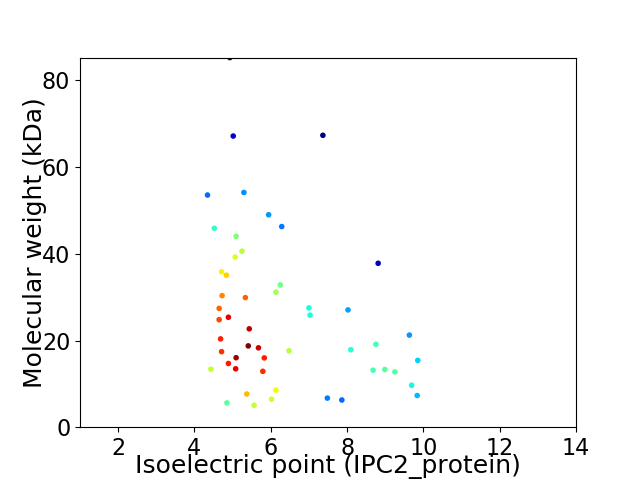
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1V0E628|A0A1V0E628_9CAUD Uncharacterized protein OS=Streptomyces phage ToastyFinz OX=1965452 GN=SEA_TOASTYFINZ_7 PE=4 SV=1
MM1 pKa = 7.72LPEE4 pKa = 5.33GIPTVRR10 pKa = 11.84VTGRR14 pKa = 11.84FLTPEE19 pKa = 4.61GKK21 pKa = 9.35PLAGQVIFRR30 pKa = 11.84APGMVTFGEE39 pKa = 4.16FDD41 pKa = 3.9VILGGPVAAPLDD53 pKa = 3.69STGAFEE59 pKa = 4.74VVLPATDD66 pKa = 3.86APGMIPTDD74 pKa = 2.84WSYY77 pKa = 12.04AVAEE81 pKa = 4.18QLAGVPMNRR90 pKa = 11.84TYY92 pKa = 10.81QVLLPAEE99 pKa = 4.44TPAVDD104 pKa = 4.44LADD107 pKa = 4.11IAPTDD112 pKa = 4.16PSTPTYY118 pKa = 9.92VAVRR122 pKa = 11.84GDD124 pKa = 3.27SAYY127 pKa = 10.19EE128 pKa = 3.88VAVEE132 pKa = 3.97AGFVGTVEE140 pKa = 3.69QWLASLIGPQGVKK153 pKa = 10.5GDD155 pKa = 3.83TGQTGAAGDD164 pKa = 3.9DD165 pKa = 3.75AYY167 pKa = 10.42EE168 pKa = 4.14VAVAAGFVGDD178 pKa = 3.97RR179 pKa = 11.84AAWLASLVGPRR190 pKa = 11.84GATGATGEE198 pKa = 4.21QGPPGTNGADD208 pKa = 3.86GADD211 pKa = 3.72GAPGVVQSVNGQSLAAVVLDD231 pKa = 4.06AADD234 pKa = 3.52VGAVPDD240 pKa = 4.27TAPGAPNGVAQLDD253 pKa = 3.96AAGKK257 pKa = 10.07VPAEE261 pKa = 4.02QLPTGTGGGAVDD273 pKa = 4.13SVNGEE278 pKa = 4.12TGVVVLDD285 pKa = 4.15AADD288 pKa = 3.76VGASPTGHH296 pKa = 6.04THH298 pKa = 5.15TAAAVGALATTARR311 pKa = 11.84GAANGVASLDD321 pKa = 3.24ASTRR325 pKa = 11.84VPIAQLPAAAGRR337 pKa = 11.84NMWTPQALGFAAWSCDD353 pKa = 3.33PYY355 pKa = 11.15TVANPVAKK363 pKa = 10.04YY364 pKa = 9.73LKK366 pKa = 8.5PQRR369 pKa = 11.84LFFVGFNITEE379 pKa = 4.23TTTVNRR385 pKa = 11.84LVMFARR391 pKa = 11.84GYY393 pKa = 11.17GGVSTNRR400 pKa = 11.84YY401 pKa = 8.59RR402 pKa = 11.84GAIYY406 pKa = 10.1RR407 pKa = 11.84DD408 pKa = 3.24TGAKK412 pKa = 9.18VVEE415 pKa = 4.48SAGVALTMAGQEE427 pKa = 4.21AGSMPAMVDD436 pKa = 3.1NHH438 pKa = 6.54IGAVPLTIASTSLAPGRR455 pKa = 11.84YY456 pKa = 5.14WAAWSLVTGGTADD469 pKa = 3.36FAFYY473 pKa = 10.21HH474 pKa = 5.52VQNEE478 pKa = 4.44APVATANFWMPGSPFARR495 pKa = 11.84AWYY498 pKa = 9.06TEE500 pKa = 3.75GQTNAALPATVSQTAAGALADD521 pKa = 3.77HH522 pKa = 7.89DD523 pKa = 4.32IPIMALANVV532 pKa = 3.79
MM1 pKa = 7.72LPEE4 pKa = 5.33GIPTVRR10 pKa = 11.84VTGRR14 pKa = 11.84FLTPEE19 pKa = 4.61GKK21 pKa = 9.35PLAGQVIFRR30 pKa = 11.84APGMVTFGEE39 pKa = 4.16FDD41 pKa = 3.9VILGGPVAAPLDD53 pKa = 3.69STGAFEE59 pKa = 4.74VVLPATDD66 pKa = 3.86APGMIPTDD74 pKa = 2.84WSYY77 pKa = 12.04AVAEE81 pKa = 4.18QLAGVPMNRR90 pKa = 11.84TYY92 pKa = 10.81QVLLPAEE99 pKa = 4.44TPAVDD104 pKa = 4.44LADD107 pKa = 4.11IAPTDD112 pKa = 4.16PSTPTYY118 pKa = 9.92VAVRR122 pKa = 11.84GDD124 pKa = 3.27SAYY127 pKa = 10.19EE128 pKa = 3.88VAVEE132 pKa = 3.97AGFVGTVEE140 pKa = 3.69QWLASLIGPQGVKK153 pKa = 10.5GDD155 pKa = 3.83TGQTGAAGDD164 pKa = 3.9DD165 pKa = 3.75AYY167 pKa = 10.42EE168 pKa = 4.14VAVAAGFVGDD178 pKa = 3.97RR179 pKa = 11.84AAWLASLVGPRR190 pKa = 11.84GATGATGEE198 pKa = 4.21QGPPGTNGADD208 pKa = 3.86GADD211 pKa = 3.72GAPGVVQSVNGQSLAAVVLDD231 pKa = 4.06AADD234 pKa = 3.52VGAVPDD240 pKa = 4.27TAPGAPNGVAQLDD253 pKa = 3.96AAGKK257 pKa = 10.07VPAEE261 pKa = 4.02QLPTGTGGGAVDD273 pKa = 4.13SVNGEE278 pKa = 4.12TGVVVLDD285 pKa = 4.15AADD288 pKa = 3.76VGASPTGHH296 pKa = 6.04THH298 pKa = 5.15TAAAVGALATTARR311 pKa = 11.84GAANGVASLDD321 pKa = 3.24ASTRR325 pKa = 11.84VPIAQLPAAAGRR337 pKa = 11.84NMWTPQALGFAAWSCDD353 pKa = 3.33PYY355 pKa = 11.15TVANPVAKK363 pKa = 10.04YY364 pKa = 9.73LKK366 pKa = 8.5PQRR369 pKa = 11.84LFFVGFNITEE379 pKa = 4.23TTTVNRR385 pKa = 11.84LVMFARR391 pKa = 11.84GYY393 pKa = 11.17GGVSTNRR400 pKa = 11.84YY401 pKa = 8.59RR402 pKa = 11.84GAIYY406 pKa = 10.1RR407 pKa = 11.84DD408 pKa = 3.24TGAKK412 pKa = 9.18VVEE415 pKa = 4.48SAGVALTMAGQEE427 pKa = 4.21AGSMPAMVDD436 pKa = 3.1NHH438 pKa = 6.54IGAVPLTIASTSLAPGRR455 pKa = 11.84YY456 pKa = 5.14WAAWSLVTGGTADD469 pKa = 3.36FAFYY473 pKa = 10.21HH474 pKa = 5.52VQNEE478 pKa = 4.44APVATANFWMPGSPFARR495 pKa = 11.84AWYY498 pKa = 9.06TEE500 pKa = 3.75GQTNAALPATVSQTAAGALADD521 pKa = 3.77HH522 pKa = 7.89DD523 pKa = 4.32IPIMALANVV532 pKa = 3.79
Molecular weight: 53.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0E690|A0A1V0E690_9CAUD Uncharacterized protein OS=Streptomyces phage ToastyFinz OX=1965452 GN=SEA_TOASTYFINZ_51 PE=4 SV=1
MM1 pKa = 7.16ATEE4 pKa = 4.41KK5 pKa = 10.54RR6 pKa = 11.84NQKK9 pKa = 7.3TAPPKK14 pKa = 9.84PGTRR18 pKa = 11.84PSIRR22 pKa = 11.84VDD24 pKa = 3.39DD25 pKa = 4.07QLAADD30 pKa = 4.7LAVVMRR36 pKa = 11.84TDD38 pKa = 3.35VNLSDD43 pKa = 3.46AVRR46 pKa = 11.84RR47 pKa = 11.84AVRR50 pKa = 11.84QLADD54 pKa = 3.32MYY56 pKa = 9.47RR57 pKa = 11.84TAWAEE62 pKa = 3.77GVVAVGTAPTLLAYY76 pKa = 9.94QLQQDD81 pKa = 4.24PAMLPPRR88 pKa = 11.84PTPAPAPTSAYY99 pKa = 9.88DD100 pKa = 3.88ARR102 pKa = 11.84PQPSTPPVGHH112 pKa = 7.14PAAPVARR119 pKa = 11.84PWAPAPGRR127 pKa = 11.84GPQVTRR133 pKa = 11.84SGPFPGVPVRR143 pKa = 11.84RR144 pKa = 11.84PP145 pKa = 3.01
MM1 pKa = 7.16ATEE4 pKa = 4.41KK5 pKa = 10.54RR6 pKa = 11.84NQKK9 pKa = 7.3TAPPKK14 pKa = 9.84PGTRR18 pKa = 11.84PSIRR22 pKa = 11.84VDD24 pKa = 3.39DD25 pKa = 4.07QLAADD30 pKa = 4.7LAVVMRR36 pKa = 11.84TDD38 pKa = 3.35VNLSDD43 pKa = 3.46AVRR46 pKa = 11.84RR47 pKa = 11.84AVRR50 pKa = 11.84QLADD54 pKa = 3.32MYY56 pKa = 9.47RR57 pKa = 11.84TAWAEE62 pKa = 3.77GVVAVGTAPTLLAYY76 pKa = 9.94QLQQDD81 pKa = 4.24PAMLPPRR88 pKa = 11.84PTPAPAPTSAYY99 pKa = 9.88DD100 pKa = 3.88ARR102 pKa = 11.84PQPSTPPVGHH112 pKa = 7.14PAAPVARR119 pKa = 11.84PWAPAPGRR127 pKa = 11.84GPQVTRR133 pKa = 11.84SGPFPGVPVRR143 pKa = 11.84RR144 pKa = 11.84PP145 pKa = 3.01
Molecular weight: 15.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12672 |
49 |
780 |
243.7 |
26.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.885 ± 0.655 | 0.679 ± 0.116 |
6.7 ± 0.312 | 5.997 ± 0.323 |
1.918 ± 0.148 | 8.546 ± 0.513 |
2.131 ± 0.178 | 3.054 ± 0.223 |
1.989 ± 0.24 | 8.365 ± 0.406 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.705 ± 0.106 | 2.107 ± 0.176 |
6.763 ± 0.407 | 3.843 ± 0.184 |
8.909 ± 0.494 | 3.969 ± 0.199 |
6.282 ± 0.28 | 7.268 ± 0.325 |
1.87 ± 0.125 | 2.02 ± 0.17 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
