
Human feces smacovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus humas3
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
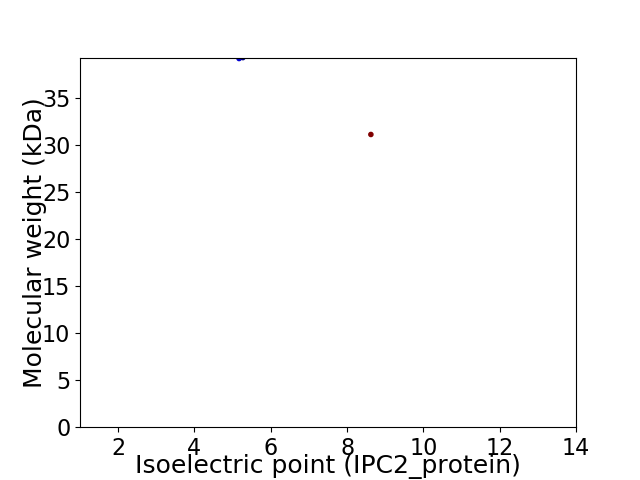
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A142J7I4|A0A142J7I4_9VIRU Replication associated protein OS=Human feces smacovirus 3 OX=1820159 PE=4 SV=1
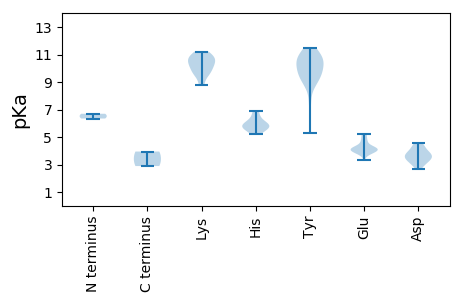
MM1 pKa = 6.69ATNYY5 pKa = 10.66ASASYY10 pKa = 10.76QEE12 pKa = 4.29ILDD15 pKa = 3.88VNTVKK20 pKa = 11.05GNVTIIGIHH29 pKa = 5.68TPTGNTPVRR38 pKa = 11.84RR39 pKa = 11.84LSGFFSQFRR48 pKa = 11.84KK49 pKa = 9.6FRR51 pKa = 11.84YY52 pKa = 9.1KK53 pKa = 9.37GCSVQVVPAATLPADD68 pKa = 3.8PLQVGFEE75 pKa = 4.13AGEE78 pKa = 3.95NTIDD82 pKa = 3.9MRR84 pKa = 11.84DD85 pKa = 3.58MLNPILFHH93 pKa = 5.87GTHH96 pKa = 6.44GEE98 pKa = 4.08SLQIAWNNIFYY109 pKa = 10.93NHH111 pKa = 6.87DD112 pKa = 3.77RR113 pKa = 11.84FNSTDD118 pKa = 3.39TGSTGYY124 pKa = 10.12IEE126 pKa = 5.14GAGFDD131 pKa = 3.68ADD133 pKa = 4.39DD134 pKa = 3.81VNFANSTNSPVEE146 pKa = 3.83TQYY149 pKa = 11.47YY150 pKa = 8.64QAITDD155 pKa = 3.78RR156 pKa = 11.84TWRR159 pKa = 11.84KK160 pKa = 9.59FGIQSTFKK168 pKa = 10.76LPFMSPRR175 pKa = 11.84VWKK178 pKa = 10.49ASTVYY183 pKa = 10.42PILPNAYY190 pKa = 9.13QDD192 pKa = 3.57QGAMNTDD199 pKa = 3.06SFMGGEE205 pKa = 4.17SALPQGTFFEE215 pKa = 5.12PSLSRR220 pKa = 11.84RR221 pKa = 11.84VGAQPTEE228 pKa = 4.05ILSQHH233 pKa = 5.25WMSSGTTPLGWLPTVTMPVNSGSVYY258 pKa = 9.93PAPPTILPKK267 pKa = 10.13IFMGILVLPPSYY279 pKa = 10.47LQEE282 pKa = 3.97LYY284 pKa = 10.53FRR286 pKa = 11.84VIVTHH291 pKa = 5.85YY292 pKa = 10.86YY293 pKa = 9.28EE294 pKa = 4.74FKK296 pKa = 11.15GFTASLAPASQMPSNIGTYY315 pKa = 10.94DD316 pKa = 3.47NAVKK320 pKa = 8.78QTATQGAKK328 pKa = 9.63EE329 pKa = 4.03AMAIDD334 pKa = 4.19EE335 pKa = 5.22PEE337 pKa = 4.64DD338 pKa = 3.54STLEE342 pKa = 4.12SPTANILRR350 pKa = 11.84VSDD353 pKa = 4.04GVFF356 pKa = 2.89
MM1 pKa = 6.69ATNYY5 pKa = 10.66ASASYY10 pKa = 10.76QEE12 pKa = 4.29ILDD15 pKa = 3.88VNTVKK20 pKa = 11.05GNVTIIGIHH29 pKa = 5.68TPTGNTPVRR38 pKa = 11.84RR39 pKa = 11.84LSGFFSQFRR48 pKa = 11.84KK49 pKa = 9.6FRR51 pKa = 11.84YY52 pKa = 9.1KK53 pKa = 9.37GCSVQVVPAATLPADD68 pKa = 3.8PLQVGFEE75 pKa = 4.13AGEE78 pKa = 3.95NTIDD82 pKa = 3.9MRR84 pKa = 11.84DD85 pKa = 3.58MLNPILFHH93 pKa = 5.87GTHH96 pKa = 6.44GEE98 pKa = 4.08SLQIAWNNIFYY109 pKa = 10.93NHH111 pKa = 6.87DD112 pKa = 3.77RR113 pKa = 11.84FNSTDD118 pKa = 3.39TGSTGYY124 pKa = 10.12IEE126 pKa = 5.14GAGFDD131 pKa = 3.68ADD133 pKa = 4.39DD134 pKa = 3.81VNFANSTNSPVEE146 pKa = 3.83TQYY149 pKa = 11.47YY150 pKa = 8.64QAITDD155 pKa = 3.78RR156 pKa = 11.84TWRR159 pKa = 11.84KK160 pKa = 9.59FGIQSTFKK168 pKa = 10.76LPFMSPRR175 pKa = 11.84VWKK178 pKa = 10.49ASTVYY183 pKa = 10.42PILPNAYY190 pKa = 9.13QDD192 pKa = 3.57QGAMNTDD199 pKa = 3.06SFMGGEE205 pKa = 4.17SALPQGTFFEE215 pKa = 5.12PSLSRR220 pKa = 11.84RR221 pKa = 11.84VGAQPTEE228 pKa = 4.05ILSQHH233 pKa = 5.25WMSSGTTPLGWLPTVTMPVNSGSVYY258 pKa = 9.93PAPPTILPKK267 pKa = 10.13IFMGILVLPPSYY279 pKa = 10.47LQEE282 pKa = 3.97LYY284 pKa = 10.53FRR286 pKa = 11.84VIVTHH291 pKa = 5.85YY292 pKa = 10.86YY293 pKa = 9.28EE294 pKa = 4.74FKK296 pKa = 11.15GFTASLAPASQMPSNIGTYY315 pKa = 10.94DD316 pKa = 3.47NAVKK320 pKa = 8.78QTATQGAKK328 pKa = 9.63EE329 pKa = 4.03AMAIDD334 pKa = 4.19EE335 pKa = 5.22PEE337 pKa = 4.64DD338 pKa = 3.54STLEE342 pKa = 4.12SPTANILRR350 pKa = 11.84VSDD353 pKa = 4.04GVFF356 pKa = 2.89
Molecular weight: 39.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142J7I4|A0A142J7I4_9VIRU Replication associated protein OS=Human feces smacovirus 3 OX=1820159 PE=4 SV=1
MM1 pKa = 6.35VQAWMITAPRR11 pKa = 11.84THH13 pKa = 5.99VSKK16 pKa = 10.63RR17 pKa = 11.84AIRR20 pKa = 11.84IMIDD24 pKa = 3.07TNDD27 pKa = 3.48CKK29 pKa = 10.87KK30 pKa = 10.1WIVAKK35 pKa = 10.47EE36 pKa = 3.87RR37 pKa = 11.84GKK39 pKa = 10.41NGYY42 pKa = 7.35EE43 pKa = 3.32HH44 pKa = 5.57WQIRR48 pKa = 11.84IEE50 pKa = 4.12SSNTGFFEE58 pKa = 3.88WCKK61 pKa = 10.68LYY63 pKa = 10.51IPTAHH68 pKa = 6.08VEE70 pKa = 3.9KK71 pKa = 10.78AEE73 pKa = 4.15RR74 pKa = 11.84GVDD77 pKa = 3.37EE78 pKa = 4.63SRR80 pKa = 11.84YY81 pKa = 5.33EE82 pKa = 4.23TKK84 pKa = 10.2EE85 pKa = 3.83GQYY88 pKa = 11.09VLYY91 pKa = 9.11TDD93 pKa = 3.29RR94 pKa = 11.84VEE96 pKa = 4.03ILKK99 pKa = 9.93QRR101 pKa = 11.84FGKK104 pKa = 8.87MRR106 pKa = 11.84PNQIRR111 pKa = 11.84ALEE114 pKa = 4.21ALEE117 pKa = 4.15STNDD121 pKa = 3.4RR122 pKa = 11.84EE123 pKa = 4.26VLVWYY128 pKa = 10.07DD129 pKa = 3.09EE130 pKa = 4.29GGNVGKK136 pKa = 10.39SWLTGALWEE145 pKa = 4.84RR146 pKa = 11.84GLAYY150 pKa = 9.61YY151 pKa = 9.54VPPTVDD157 pKa = 2.8SVKK160 pKa = 11.22AMVQWVASCYY170 pKa = 9.73QSGGWRR176 pKa = 11.84PYY178 pKa = 10.81VIIDD182 pKa = 4.59IPRR185 pKa = 11.84SWKK188 pKa = 9.59WSEE191 pKa = 3.76QLYY194 pKa = 10.02VAIEE198 pKa = 4.26SIKK201 pKa = 11.03DD202 pKa = 3.15GLVYY206 pKa = 9.39DD207 pKa = 4.17TRR209 pKa = 11.84YY210 pKa = 9.55HH211 pKa = 5.6AQCINIRR218 pKa = 11.84GVKK221 pKa = 10.09VLVLTNTQPKK231 pKa = 9.97LDD233 pKa = 4.58KK234 pKa = 10.73LSQDD238 pKa = 2.67RR239 pKa = 11.84WRR241 pKa = 11.84ICVFRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84GSALIVTLWADD260 pKa = 4.28FYY262 pKa = 11.04PPLGGG267 pKa = 3.95
MM1 pKa = 6.35VQAWMITAPRR11 pKa = 11.84THH13 pKa = 5.99VSKK16 pKa = 10.63RR17 pKa = 11.84AIRR20 pKa = 11.84IMIDD24 pKa = 3.07TNDD27 pKa = 3.48CKK29 pKa = 10.87KK30 pKa = 10.1WIVAKK35 pKa = 10.47EE36 pKa = 3.87RR37 pKa = 11.84GKK39 pKa = 10.41NGYY42 pKa = 7.35EE43 pKa = 3.32HH44 pKa = 5.57WQIRR48 pKa = 11.84IEE50 pKa = 4.12SSNTGFFEE58 pKa = 3.88WCKK61 pKa = 10.68LYY63 pKa = 10.51IPTAHH68 pKa = 6.08VEE70 pKa = 3.9KK71 pKa = 10.78AEE73 pKa = 4.15RR74 pKa = 11.84GVDD77 pKa = 3.37EE78 pKa = 4.63SRR80 pKa = 11.84YY81 pKa = 5.33EE82 pKa = 4.23TKK84 pKa = 10.2EE85 pKa = 3.83GQYY88 pKa = 11.09VLYY91 pKa = 9.11TDD93 pKa = 3.29RR94 pKa = 11.84VEE96 pKa = 4.03ILKK99 pKa = 9.93QRR101 pKa = 11.84FGKK104 pKa = 8.87MRR106 pKa = 11.84PNQIRR111 pKa = 11.84ALEE114 pKa = 4.21ALEE117 pKa = 4.15STNDD121 pKa = 3.4RR122 pKa = 11.84EE123 pKa = 4.26VLVWYY128 pKa = 10.07DD129 pKa = 3.09EE130 pKa = 4.29GGNVGKK136 pKa = 10.39SWLTGALWEE145 pKa = 4.84RR146 pKa = 11.84GLAYY150 pKa = 9.61YY151 pKa = 9.54VPPTVDD157 pKa = 2.8SVKK160 pKa = 11.22AMVQWVASCYY170 pKa = 9.73QSGGWRR176 pKa = 11.84PYY178 pKa = 10.81VIIDD182 pKa = 4.59IPRR185 pKa = 11.84SWKK188 pKa = 9.59WSEE191 pKa = 3.76QLYY194 pKa = 10.02VAIEE198 pKa = 4.26SIKK201 pKa = 11.03DD202 pKa = 3.15GLVYY206 pKa = 9.39DD207 pKa = 4.17TRR209 pKa = 11.84YY210 pKa = 9.55HH211 pKa = 5.6AQCINIRR218 pKa = 11.84GVKK221 pKa = 10.09VLVLTNTQPKK231 pKa = 9.97LDD233 pKa = 4.58KK234 pKa = 10.73LSQDD238 pKa = 2.67RR239 pKa = 11.84WRR241 pKa = 11.84ICVFRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84GSALIVTLWADD260 pKa = 4.28FYY262 pKa = 11.04PPLGGG267 pKa = 3.95
Molecular weight: 31.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
623 |
267 |
356 |
311.5 |
35.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.902 ± 0.538 | 0.963 ± 0.538 |
4.655 ± 0.127 | 5.136 ± 0.728 |
4.173 ± 1.36 | 7.384 ± 0.158 |
1.605 ± 0.063 | 6.26 ± 0.506 |
4.334 ± 1.202 | 6.421 ± 0.19 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.411 | 4.334 ± 0.791 |
5.939 ± 1.297 | 4.494 ± 0.221 |
5.618 ± 1.55 | 6.902 ± 0.981 |
7.544 ± 1.36 | 7.223 ± 0.822 |
2.889 ± 1.17 | 4.655 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
