
Chelonia mydas papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyozetapapillomavirus; Dyozetapapillomavirus 1
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
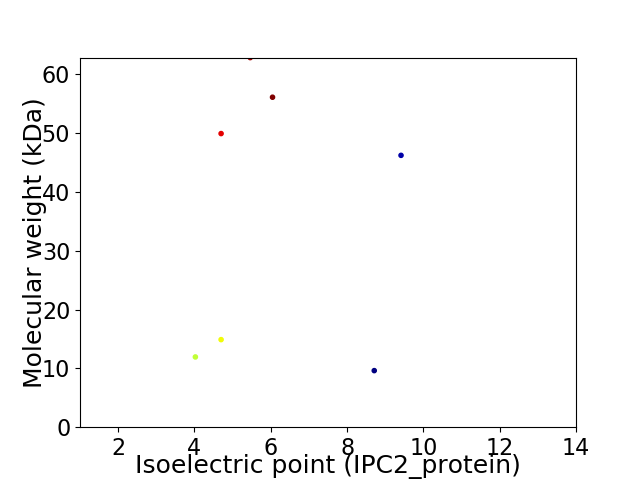
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
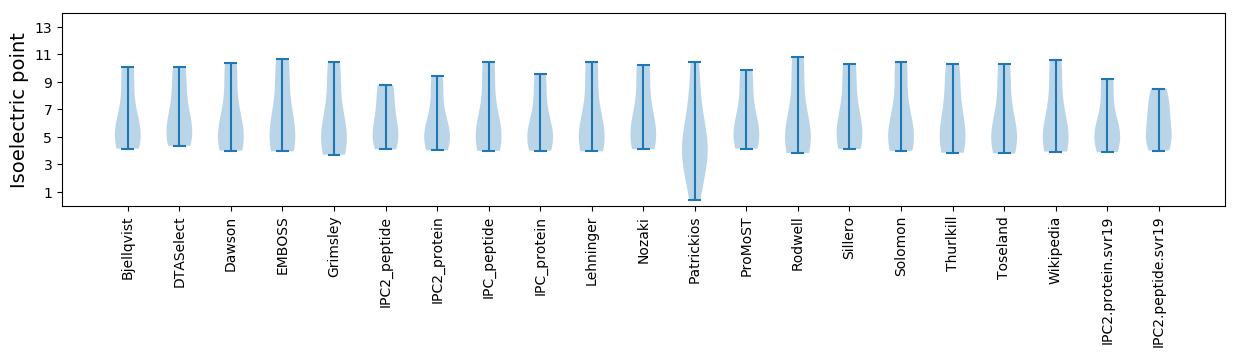
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6RUP3|B6RUP3_9PAPI Replication protein OS=Chelonia mydas papillomavirus 1 OX=485242 GN=E1 PE=4 SV=1
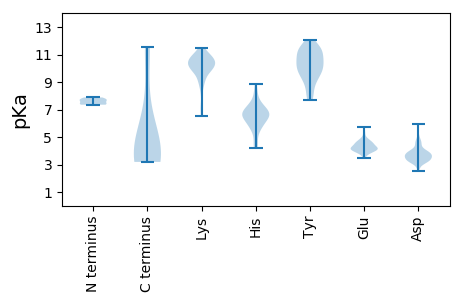
MM1 pKa = 7.71HH2 pKa = 8.17DD3 pKa = 3.85KK4 pKa = 10.76LQPQKK9 pKa = 11.01GCLSRR14 pKa = 11.84YY15 pKa = 9.07ICACCRR21 pKa = 11.84QEE23 pKa = 4.6VDD25 pKa = 3.97FAQQEE30 pKa = 4.63VCVMLSDD37 pKa = 3.68QLGMLVCRR45 pKa = 11.84SCEE48 pKa = 3.93QSLLPHH54 pKa = 6.6EE55 pKa = 4.88LSDD58 pKa = 5.05AISHH62 pKa = 6.49ACMTEE67 pKa = 3.9VSVSAGFDD75 pKa = 3.37LHH77 pKa = 8.86SDD79 pKa = 3.76DD80 pKa = 5.95SDD82 pKa = 3.65LASYY86 pKa = 11.33SDD88 pKa = 3.23ISDD91 pKa = 3.36WDD93 pKa = 3.92SEE95 pKa = 4.33EE96 pKa = 5.67SPDD99 pKa = 4.08TDD101 pKa = 3.2TDD103 pKa = 3.97FSGSHH108 pKa = 5.86
MM1 pKa = 7.71HH2 pKa = 8.17DD3 pKa = 3.85KK4 pKa = 10.76LQPQKK9 pKa = 11.01GCLSRR14 pKa = 11.84YY15 pKa = 9.07ICACCRR21 pKa = 11.84QEE23 pKa = 4.6VDD25 pKa = 3.97FAQQEE30 pKa = 4.63VCVMLSDD37 pKa = 3.68QLGMLVCRR45 pKa = 11.84SCEE48 pKa = 3.93QSLLPHH54 pKa = 6.6EE55 pKa = 4.88LSDD58 pKa = 5.05AISHH62 pKa = 6.49ACMTEE67 pKa = 3.9VSVSAGFDD75 pKa = 3.37LHH77 pKa = 8.86SDD79 pKa = 3.76DD80 pKa = 5.95SDD82 pKa = 3.65LASYY86 pKa = 11.33SDD88 pKa = 3.23ISDD91 pKa = 3.36WDD93 pKa = 3.92SEE95 pKa = 4.33EE96 pKa = 5.67SPDD99 pKa = 4.08TDD101 pKa = 3.2TDD103 pKa = 3.97FSGSHH108 pKa = 5.86
Molecular weight: 11.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6RUP5|B6RUP5_9PAPI Putative E4 protein OS=Chelonia mydas papillomavirus 1 OX=485242 GN=E4 PE=4 SV=1
MM1 pKa = 7.42MDD3 pKa = 3.4ANQRR7 pKa = 11.84LMEE10 pKa = 4.12VQNIQLSIIEE20 pKa = 4.32QDD22 pKa = 3.19SHH24 pKa = 5.6TLGSILEE31 pKa = 4.58FYY33 pKa = 10.93KK34 pKa = 10.93AMKK37 pKa = 10.32VEE39 pKa = 3.98YY40 pKa = 9.96LLLAAARR47 pKa = 11.84KK48 pKa = 9.53KK49 pKa = 10.43GRR51 pKa = 11.84LHH53 pKa = 7.37IGVQRR58 pKa = 11.84VPPMQVSEE66 pKa = 4.3TKK68 pKa = 10.33YY69 pKa = 10.38RR70 pKa = 11.84EE71 pKa = 3.92ASTMIVLIEE80 pKa = 4.59SLMQSQFRR88 pKa = 11.84DD89 pKa = 3.27RR90 pKa = 11.84KK91 pKa = 9.66FSLHH95 pKa = 5.72EE96 pKa = 3.95LQYY99 pKa = 10.61SLVMTPPEE107 pKa = 4.1YY108 pKa = 9.85TVKK111 pKa = 10.48QGPKK115 pKa = 9.23QVYY118 pKa = 7.66ITYY121 pKa = 9.99SDD123 pKa = 3.95PSQTTEE129 pKa = 5.15QFTKK133 pKa = 9.77WKK135 pKa = 8.55TILYY139 pKa = 7.98QTDD142 pKa = 3.38DD143 pKa = 3.17WSEE146 pKa = 4.12RR147 pKa = 11.84PSLPGTHH154 pKa = 6.79DD155 pKa = 3.8PKK157 pKa = 10.67WFLAHH162 pKa = 6.76TLTDD166 pKa = 3.51KK167 pKa = 11.42NGLFIVDD174 pKa = 3.51KK175 pKa = 11.34SGDD178 pKa = 3.19KK179 pKa = 10.46DD180 pKa = 3.7YY181 pKa = 11.81YY182 pKa = 11.5AFFSTKK188 pKa = 9.65EE189 pKa = 3.85PSSAVARR196 pKa = 11.84GQWTISSTVPAAAALSRR213 pKa = 11.84QPSPRR218 pKa = 11.84RR219 pKa = 11.84SNSPNLDD226 pKa = 3.13PCTRR230 pKa = 11.84SRR232 pKa = 11.84GSHH235 pKa = 6.37CGGVSPSNPDD245 pKa = 3.35TPDD248 pKa = 3.19TAGVRR253 pKa = 11.84PRR255 pKa = 11.84DD256 pKa = 3.57RR257 pKa = 11.84TGGLGTPPKK266 pKa = 9.77SARR269 pKa = 11.84GGGGRR274 pKa = 11.84GQSTGGSKK282 pKa = 10.19LQRR285 pKa = 11.84GPRR288 pKa = 11.84VTPRR292 pKa = 11.84SSRR295 pKa = 11.84QSKK298 pKa = 9.2EE299 pKa = 3.86KK300 pKa = 11.06NPVSPAEE307 pKa = 3.88VGQNRR312 pKa = 11.84KK313 pKa = 6.56TVSGPGTRR321 pKa = 11.84LDD323 pKa = 3.51RR324 pKa = 11.84LIRR327 pKa = 11.84EE328 pKa = 4.32ARR330 pKa = 11.84DD331 pKa = 3.33PPGVIIEE338 pKa = 4.75GSTSQIKK345 pKa = 9.77HH346 pKa = 4.82LRR348 pKa = 11.84RR349 pKa = 11.84RR350 pKa = 11.84IQLGPLKK357 pKa = 9.73YY358 pKa = 10.59LRR360 pKa = 11.84VTSTWHH366 pKa = 5.73WIVNKK371 pKa = 10.07KK372 pKa = 7.2VQKK375 pKa = 7.36PCKK378 pKa = 8.9MIVVFSNNAEE388 pKa = 3.98RR389 pKa = 11.84QTFLHH394 pKa = 6.71LFRR397 pKa = 11.84VEE399 pKa = 3.76GDD401 pKa = 3.82GISVRR406 pKa = 11.84LCSFNGLL413 pKa = 3.61
MM1 pKa = 7.42MDD3 pKa = 3.4ANQRR7 pKa = 11.84LMEE10 pKa = 4.12VQNIQLSIIEE20 pKa = 4.32QDD22 pKa = 3.19SHH24 pKa = 5.6TLGSILEE31 pKa = 4.58FYY33 pKa = 10.93KK34 pKa = 10.93AMKK37 pKa = 10.32VEE39 pKa = 3.98YY40 pKa = 9.96LLLAAARR47 pKa = 11.84KK48 pKa = 9.53KK49 pKa = 10.43GRR51 pKa = 11.84LHH53 pKa = 7.37IGVQRR58 pKa = 11.84VPPMQVSEE66 pKa = 4.3TKK68 pKa = 10.33YY69 pKa = 10.38RR70 pKa = 11.84EE71 pKa = 3.92ASTMIVLIEE80 pKa = 4.59SLMQSQFRR88 pKa = 11.84DD89 pKa = 3.27RR90 pKa = 11.84KK91 pKa = 9.66FSLHH95 pKa = 5.72EE96 pKa = 3.95LQYY99 pKa = 10.61SLVMTPPEE107 pKa = 4.1YY108 pKa = 9.85TVKK111 pKa = 10.48QGPKK115 pKa = 9.23QVYY118 pKa = 7.66ITYY121 pKa = 9.99SDD123 pKa = 3.95PSQTTEE129 pKa = 5.15QFTKK133 pKa = 9.77WKK135 pKa = 8.55TILYY139 pKa = 7.98QTDD142 pKa = 3.38DD143 pKa = 3.17WSEE146 pKa = 4.12RR147 pKa = 11.84PSLPGTHH154 pKa = 6.79DD155 pKa = 3.8PKK157 pKa = 10.67WFLAHH162 pKa = 6.76TLTDD166 pKa = 3.51KK167 pKa = 11.42NGLFIVDD174 pKa = 3.51KK175 pKa = 11.34SGDD178 pKa = 3.19KK179 pKa = 10.46DD180 pKa = 3.7YY181 pKa = 11.81YY182 pKa = 11.5AFFSTKK188 pKa = 9.65EE189 pKa = 3.85PSSAVARR196 pKa = 11.84GQWTISSTVPAAAALSRR213 pKa = 11.84QPSPRR218 pKa = 11.84RR219 pKa = 11.84SNSPNLDD226 pKa = 3.13PCTRR230 pKa = 11.84SRR232 pKa = 11.84GSHH235 pKa = 6.37CGGVSPSNPDD245 pKa = 3.35TPDD248 pKa = 3.19TAGVRR253 pKa = 11.84PRR255 pKa = 11.84DD256 pKa = 3.57RR257 pKa = 11.84TGGLGTPPKK266 pKa = 9.77SARR269 pKa = 11.84GGGGRR274 pKa = 11.84GQSTGGSKK282 pKa = 10.19LQRR285 pKa = 11.84GPRR288 pKa = 11.84VTPRR292 pKa = 11.84SSRR295 pKa = 11.84QSKK298 pKa = 9.2EE299 pKa = 3.86KK300 pKa = 11.06NPVSPAEE307 pKa = 3.88VGQNRR312 pKa = 11.84KK313 pKa = 6.56TVSGPGTRR321 pKa = 11.84LDD323 pKa = 3.51RR324 pKa = 11.84LIRR327 pKa = 11.84EE328 pKa = 4.32ARR330 pKa = 11.84DD331 pKa = 3.33PPGVIIEE338 pKa = 4.75GSTSQIKK345 pKa = 9.77HH346 pKa = 4.82LRR348 pKa = 11.84RR349 pKa = 11.84RR350 pKa = 11.84IQLGPLKK357 pKa = 9.73YY358 pKa = 10.59LRR360 pKa = 11.84VTSTWHH366 pKa = 5.73WIVNKK371 pKa = 10.07KK372 pKa = 7.2VQKK375 pKa = 7.36PCKK378 pKa = 8.9MIVVFSNNAEE388 pKa = 3.98RR389 pKa = 11.84QTFLHH394 pKa = 6.71LFRR397 pKa = 11.84VEE399 pKa = 3.76GDD401 pKa = 3.82GISVRR406 pKa = 11.84LCSFNGLL413 pKa = 3.61
Molecular weight: 46.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2268 |
85 |
559 |
324.0 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.876 | 2.381 ± 0.777 |
6.481 ± 0.642 | 5.159 ± 0.367 |
4.145 ± 0.594 | 6.481 ± 0.57 |
2.072 ± 0.295 | 4.409 ± 0.42 |
4.718 ± 0.677 | 8.069 ± 0.737 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.94 ± 0.246 | 3.483 ± 0.766 |
8.069 ± 1.74 | 4.056 ± 0.679 |
5.908 ± 0.643 | 8.951 ± 0.777 |
6.834 ± 0.768 | 6.217 ± 0.514 |
1.455 ± 0.246 | 2.778 ± 0.295 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
