
Ornithinibacillus halophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Ornithinibacillus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
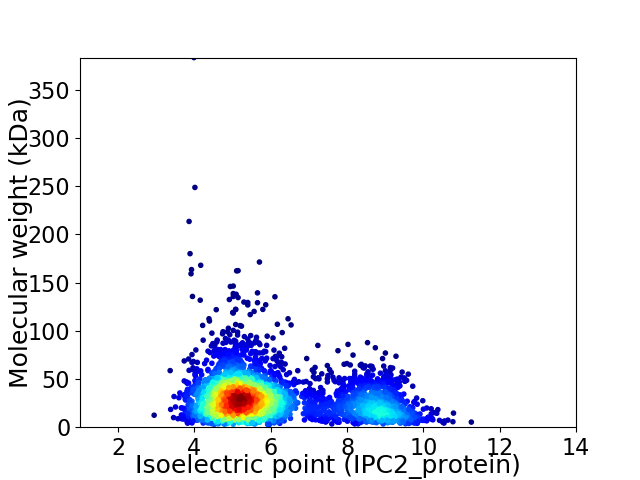
Virtual 2D-PAGE plot for 3462 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5FLS8|A0A1M5FLS8_9BACI Probable molybdenum cofactor guanylyltransferase OS=Ornithinibacillus halophilus OX=930117 GN=mobA PE=3 SV=1
MM1 pKa = 7.46ALIGLVLILAACGGDD16 pKa = 3.26KK17 pKa = 11.23ANIAVGPAGSATNTVSSLILEE38 pKa = 4.48AYY40 pKa = 9.9GIEE43 pKa = 4.22EE44 pKa = 4.02GDD46 pKa = 3.44YY47 pKa = 9.91TAFEE51 pKa = 4.46EE52 pKa = 5.22GFGDD56 pKa = 4.68AADD59 pKa = 4.27GVQDD63 pKa = 3.75GNIDD67 pKa = 3.25ISIGILGLPAGSIEE81 pKa = 4.2NLHH84 pKa = 6.78ASAGDD89 pKa = 3.39AKK91 pKa = 10.24MLSLSKK97 pKa = 10.5EE98 pKa = 3.52AVAYY102 pKa = 9.91IEE104 pKa = 4.63EE105 pKa = 4.07NSAYY109 pKa = 10.34RR110 pKa = 11.84EE111 pKa = 3.97MVIPADD117 pKa = 3.31SYY119 pKa = 11.46EE120 pKa = 3.98FLEE123 pKa = 4.42EE124 pKa = 4.6DD125 pKa = 3.77VTTVTAYY132 pKa = 10.48AILMGNTNTISEE144 pKa = 4.29EE145 pKa = 4.02LGYY148 pKa = 10.79QLAKK152 pKa = 11.08VMIEE156 pKa = 3.82NSSDD160 pKa = 2.86ISHH163 pKa = 6.63AQGAQMTLDD172 pKa = 3.31NALNGLEE179 pKa = 5.04DD180 pKa = 3.87MPIHH184 pKa = 6.75PGAARR189 pKa = 11.84YY190 pKa = 9.54YY191 pKa = 9.51EE192 pKa = 4.13EE193 pKa = 5.05QGLEE197 pKa = 3.84FDD199 pKa = 4.75NPIAEE204 pKa = 4.47LNVTEE209 pKa = 5.47DD210 pKa = 3.5VTEE213 pKa = 4.16LTLGTGSSGGTYY225 pKa = 10.06FPLGGEE231 pKa = 4.29MATIWKK237 pKa = 10.3DD238 pKa = 3.36NIEE241 pKa = 4.74GINVTSQEE249 pKa = 4.01TGASLEE255 pKa = 3.84NMARR259 pKa = 11.84IGEE262 pKa = 4.27GTMDD266 pKa = 4.28LGMAVHH272 pKa = 6.94GPAMDD277 pKa = 3.98GVNGEE282 pKa = 4.74GEE284 pKa = 3.86FDD286 pKa = 3.88APIEE290 pKa = 3.93NAAFIGHH297 pKa = 7.62IYY299 pKa = 9.97PEE301 pKa = 4.28VVQIVTRR308 pKa = 11.84EE309 pKa = 4.02STDD312 pKa = 2.82IDD314 pKa = 3.66SFEE317 pKa = 4.37DD318 pKa = 3.44LKK320 pKa = 11.61
MM1 pKa = 7.46ALIGLVLILAACGGDD16 pKa = 3.26KK17 pKa = 11.23ANIAVGPAGSATNTVSSLILEE38 pKa = 4.48AYY40 pKa = 9.9GIEE43 pKa = 4.22EE44 pKa = 4.02GDD46 pKa = 3.44YY47 pKa = 9.91TAFEE51 pKa = 4.46EE52 pKa = 5.22GFGDD56 pKa = 4.68AADD59 pKa = 4.27GVQDD63 pKa = 3.75GNIDD67 pKa = 3.25ISIGILGLPAGSIEE81 pKa = 4.2NLHH84 pKa = 6.78ASAGDD89 pKa = 3.39AKK91 pKa = 10.24MLSLSKK97 pKa = 10.5EE98 pKa = 3.52AVAYY102 pKa = 9.91IEE104 pKa = 4.63EE105 pKa = 4.07NSAYY109 pKa = 10.34RR110 pKa = 11.84EE111 pKa = 3.97MVIPADD117 pKa = 3.31SYY119 pKa = 11.46EE120 pKa = 3.98FLEE123 pKa = 4.42EE124 pKa = 4.6DD125 pKa = 3.77VTTVTAYY132 pKa = 10.48AILMGNTNTISEE144 pKa = 4.29EE145 pKa = 4.02LGYY148 pKa = 10.79QLAKK152 pKa = 11.08VMIEE156 pKa = 3.82NSSDD160 pKa = 2.86ISHH163 pKa = 6.63AQGAQMTLDD172 pKa = 3.31NALNGLEE179 pKa = 5.04DD180 pKa = 3.87MPIHH184 pKa = 6.75PGAARR189 pKa = 11.84YY190 pKa = 9.54YY191 pKa = 9.51EE192 pKa = 4.13EE193 pKa = 5.05QGLEE197 pKa = 3.84FDD199 pKa = 4.75NPIAEE204 pKa = 4.47LNVTEE209 pKa = 5.47DD210 pKa = 3.5VTEE213 pKa = 4.16LTLGTGSSGGTYY225 pKa = 10.06FPLGGEE231 pKa = 4.29MATIWKK237 pKa = 10.3DD238 pKa = 3.36NIEE241 pKa = 4.74GINVTSQEE249 pKa = 4.01TGASLEE255 pKa = 3.84NMARR259 pKa = 11.84IGEE262 pKa = 4.27GTMDD266 pKa = 4.28LGMAVHH272 pKa = 6.94GPAMDD277 pKa = 3.98GVNGEE282 pKa = 4.74GEE284 pKa = 3.86FDD286 pKa = 3.88APIEE290 pKa = 3.93NAAFIGHH297 pKa = 7.62IYY299 pKa = 9.97PEE301 pKa = 4.28VVQIVTRR308 pKa = 11.84EE309 pKa = 4.02STDD312 pKa = 2.82IDD314 pKa = 3.66SFEE317 pKa = 4.37DD318 pKa = 3.44LKK320 pKa = 11.61
Molecular weight: 33.66 kDa
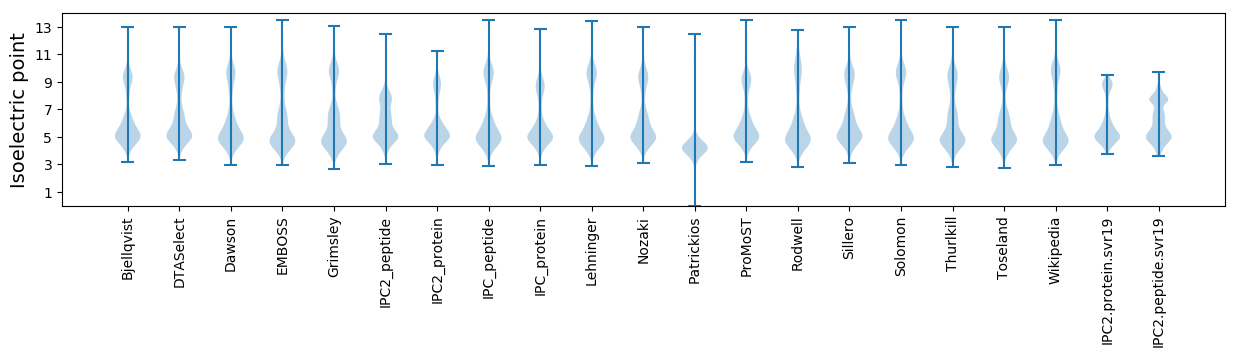
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5ME28|A0A1M5ME28_9BACI Ribosomal RNA small subunit methyltransferase A OS=Ornithinibacillus halophilus OX=930117 GN=rsmA PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
975949 |
25 |
3510 |
281.9 |
31.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.292 ± 0.049 | 0.587 ± 0.011 |
5.529 ± 0.038 | 7.938 ± 0.048 |
4.544 ± 0.034 | 6.641 ± 0.047 |
2.04 ± 0.021 | 8.283 ± 0.042 |
6.793 ± 0.045 | 9.517 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.736 ± 0.021 | 4.915 ± 0.036 |
3.508 ± 0.025 | 3.704 ± 0.033 |
3.822 ± 0.032 | 5.937 ± 0.032 |
5.451 ± 0.03 | 7.081 ± 0.038 |
1.001 ± 0.016 | 3.684 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
