
alpha proteobacterium HIMB114
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Pelagibacterales; Pelagibacteraceae; unclassified Pelagibacteraceae
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
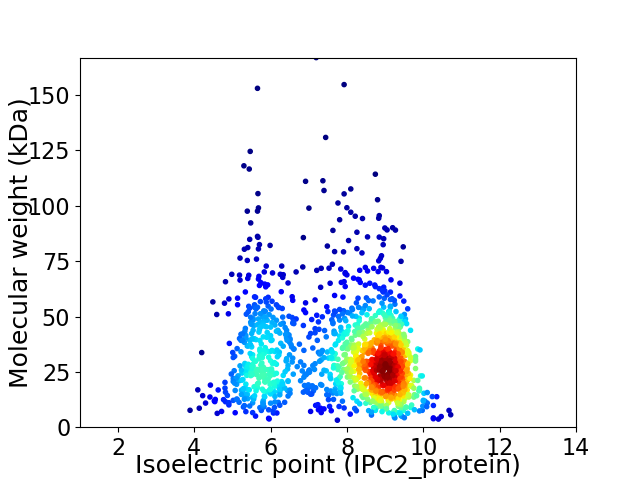
Virtual 2D-PAGE plot for 1321 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0RNL6|D0RNL6_9PROT 50S ribosomal protein L1 OS=alpha proteobacterium HIMB114 OX=684719 GN=rplA PE=3 SV=1
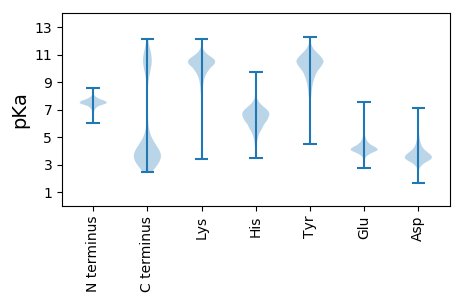
MM1 pKa = 7.59KK2 pKa = 10.46KK3 pKa = 10.35LLATTAIVSVSFASVALAEE22 pKa = 4.38VTVKK26 pKa = 10.77GSIEE30 pKa = 3.64QTINSRR36 pKa = 11.84SYY38 pKa = 9.63NKK40 pKa = 10.06AANEE44 pKa = 4.06HH45 pKa = 6.7KK46 pKa = 10.14GTGSMGQEE54 pKa = 3.93TNLTVSSSGEE64 pKa = 3.9LDD66 pKa = 2.96NGLSIKK72 pKa = 10.63GSFRR76 pKa = 11.84SEE78 pKa = 4.28DD79 pKa = 3.26GSVDD83 pKa = 3.05HH84 pKa = 7.19SSIKK88 pKa = 10.57VSGDD92 pKa = 3.1TLGFEE97 pKa = 4.51IGADD101 pKa = 3.32TGSTIHH107 pKa = 5.63TQINPTVGDD116 pKa = 3.9SVWFVTGAQGDD127 pKa = 4.15DD128 pKa = 3.26GFTTYY133 pKa = 10.12EE134 pKa = 3.85AHH136 pKa = 7.08DD137 pKa = 3.95VQHH140 pKa = 6.98IGIDD144 pKa = 3.43AKK146 pKa = 11.02LGGMTAAVNYY156 pKa = 9.97APSNNGISAGDD167 pKa = 3.85SNKK170 pKa = 9.53TDD172 pKa = 3.28AGGSATEE179 pKa = 4.01MLLKK183 pKa = 10.4GSIMEE188 pKa = 4.47GVNFLIGQEE197 pKa = 4.15KK198 pKa = 10.05IEE200 pKa = 4.21AANSGGSEE208 pKa = 4.15EE209 pKa = 4.37TEE211 pKa = 3.56KK212 pKa = 10.24TYY214 pKa = 10.7QVSYY218 pKa = 10.71SAGQFALGASIRR230 pKa = 11.84DD231 pKa = 3.66FDD233 pKa = 6.24DD234 pKa = 4.72GDD236 pKa = 3.91STATAGTTDD245 pKa = 3.29EE246 pKa = 4.38VTYY249 pKa = 9.21LTATYY254 pKa = 10.43AVNDD258 pKa = 3.81QVSLGIQSVTAEE270 pKa = 4.18SEE272 pKa = 4.42GTGTSATDD280 pKa = 3.38EE281 pKa = 4.24EE282 pKa = 4.95TLGISLGYY290 pKa = 9.76NLGPIGVEE298 pKa = 3.69VMYY301 pKa = 11.16AEE303 pKa = 4.61TDD305 pKa = 3.49NLAHH309 pKa = 6.8SNADD313 pKa = 3.39DD314 pKa = 3.9VEE316 pKa = 4.66GVQIRR321 pKa = 11.84TVYY324 pKa = 10.65KK325 pKa = 10.04FF326 pKa = 3.04
MM1 pKa = 7.59KK2 pKa = 10.46KK3 pKa = 10.35LLATTAIVSVSFASVALAEE22 pKa = 4.38VTVKK26 pKa = 10.77GSIEE30 pKa = 3.64QTINSRR36 pKa = 11.84SYY38 pKa = 9.63NKK40 pKa = 10.06AANEE44 pKa = 4.06HH45 pKa = 6.7KK46 pKa = 10.14GTGSMGQEE54 pKa = 3.93TNLTVSSSGEE64 pKa = 3.9LDD66 pKa = 2.96NGLSIKK72 pKa = 10.63GSFRR76 pKa = 11.84SEE78 pKa = 4.28DD79 pKa = 3.26GSVDD83 pKa = 3.05HH84 pKa = 7.19SSIKK88 pKa = 10.57VSGDD92 pKa = 3.1TLGFEE97 pKa = 4.51IGADD101 pKa = 3.32TGSTIHH107 pKa = 5.63TQINPTVGDD116 pKa = 3.9SVWFVTGAQGDD127 pKa = 4.15DD128 pKa = 3.26GFTTYY133 pKa = 10.12EE134 pKa = 3.85AHH136 pKa = 7.08DD137 pKa = 3.95VQHH140 pKa = 6.98IGIDD144 pKa = 3.43AKK146 pKa = 11.02LGGMTAAVNYY156 pKa = 9.97APSNNGISAGDD167 pKa = 3.85SNKK170 pKa = 9.53TDD172 pKa = 3.28AGGSATEE179 pKa = 4.01MLLKK183 pKa = 10.4GSIMEE188 pKa = 4.47GVNFLIGQEE197 pKa = 4.15KK198 pKa = 10.05IEE200 pKa = 4.21AANSGGSEE208 pKa = 4.15EE209 pKa = 4.37TEE211 pKa = 3.56KK212 pKa = 10.24TYY214 pKa = 10.7QVSYY218 pKa = 10.71SAGQFALGASIRR230 pKa = 11.84DD231 pKa = 3.66FDD233 pKa = 6.24DD234 pKa = 4.72GDD236 pKa = 3.91STATAGTTDD245 pKa = 3.29EE246 pKa = 4.38VTYY249 pKa = 9.21LTATYY254 pKa = 10.43AVNDD258 pKa = 3.81QVSLGIQSVTAEE270 pKa = 4.18SEE272 pKa = 4.42GTGTSATDD280 pKa = 3.38EE281 pKa = 4.24EE282 pKa = 4.95TLGISLGYY290 pKa = 9.76NLGPIGVEE298 pKa = 3.69VMYY301 pKa = 11.16AEE303 pKa = 4.61TDD305 pKa = 3.49NLAHH309 pKa = 6.8SNADD313 pKa = 3.39DD314 pKa = 3.9VEE316 pKa = 4.66GVQIRR321 pKa = 11.84TVYY324 pKa = 10.65KK325 pKa = 10.04FF326 pKa = 3.04
Molecular weight: 33.71 kDa
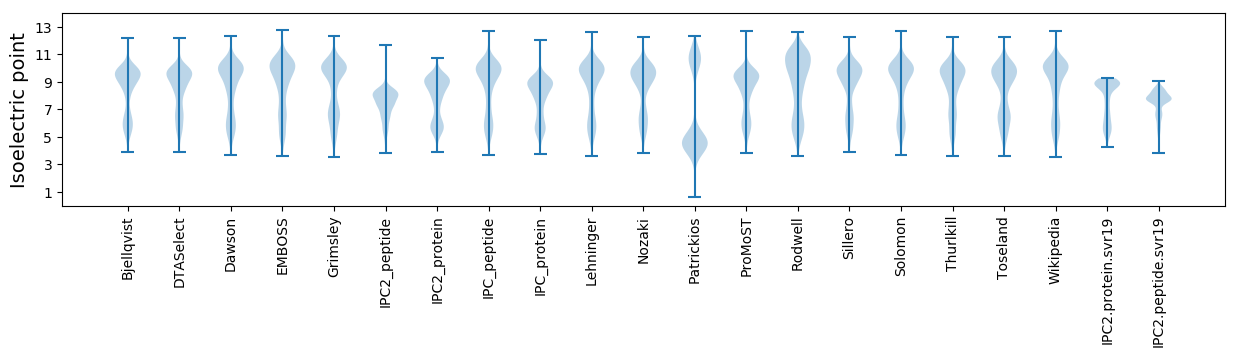
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0RNS5|D0RNS5_9PROT 5-aminolevulinate synthase OS=alpha proteobacterium HIMB114 OX=684719 GN=HIMB114_00004490 PE=3 SV=1
MM1 pKa = 6.66VTRR4 pKa = 11.84KK5 pKa = 10.28SFTRR9 pKa = 11.84PVTKK13 pKa = 10.2TSNPQPKK20 pKa = 9.66NKK22 pKa = 9.55KK23 pKa = 9.38SKK25 pKa = 10.77DD26 pKa = 3.48WIIWAAWADD35 pKa = 3.5RR36 pKa = 11.84VTFEE40 pKa = 5.24DD41 pKa = 3.24IKK43 pKa = 11.07RR44 pKa = 11.84EE45 pKa = 3.97TGRR48 pKa = 11.84TEE50 pKa = 3.94SEE52 pKa = 4.15VIKK55 pKa = 10.68IMRR58 pKa = 11.84STLKK62 pKa = 10.43PSSFRR67 pKa = 11.84LWRR70 pKa = 11.84KK71 pKa = 8.93RR72 pKa = 11.84VHH74 pKa = 5.88TKK76 pKa = 10.29SIKK79 pKa = 8.91NLKK82 pKa = 9.62KK83 pKa = 10.46FRR85 pKa = 11.84EE86 pKa = 4.24DD87 pKa = 3.02RR88 pKa = 11.84QKK90 pKa = 10.64IRR92 pKa = 11.84NKK94 pKa = 10.3SYY96 pKa = 11.29KK97 pKa = 10.37NYY99 pKa = 10.72LL100 pKa = 3.4
MM1 pKa = 6.66VTRR4 pKa = 11.84KK5 pKa = 10.28SFTRR9 pKa = 11.84PVTKK13 pKa = 10.2TSNPQPKK20 pKa = 9.66NKK22 pKa = 9.55KK23 pKa = 9.38SKK25 pKa = 10.77DD26 pKa = 3.48WIIWAAWADD35 pKa = 3.5RR36 pKa = 11.84VTFEE40 pKa = 5.24DD41 pKa = 3.24IKK43 pKa = 11.07RR44 pKa = 11.84EE45 pKa = 3.97TGRR48 pKa = 11.84TEE50 pKa = 3.94SEE52 pKa = 4.15VIKK55 pKa = 10.68IMRR58 pKa = 11.84STLKK62 pKa = 10.43PSSFRR67 pKa = 11.84LWRR70 pKa = 11.84KK71 pKa = 8.93RR72 pKa = 11.84VHH74 pKa = 5.88TKK76 pKa = 10.29SIKK79 pKa = 8.91NLKK82 pKa = 9.62KK83 pKa = 10.46FRR85 pKa = 11.84EE86 pKa = 4.24DD87 pKa = 3.02RR88 pKa = 11.84QKK90 pKa = 10.64IRR92 pKa = 11.84NKK94 pKa = 10.3SYY96 pKa = 11.29KK97 pKa = 10.37NYY99 pKa = 10.72LL100 pKa = 3.4
Molecular weight: 12.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
397694 |
30 |
1502 |
301.1 |
34.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.423 ± 0.073 | 1.03 ± 0.02 |
5.191 ± 0.056 | 5.894 ± 0.068 |
5.582 ± 0.082 | 6.13 ± 0.069 |
1.581 ± 0.03 | 9.552 ± 0.081 |
10.909 ± 0.098 | 9.445 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.032 | 6.698 ± 0.086 |
3.148 ± 0.039 | 2.839 ± 0.035 |
3.139 ± 0.048 | 6.832 ± 0.052 |
4.63 ± 0.042 | 5.652 ± 0.067 |
0.833 ± 0.024 | 3.422 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
