
Capybara microvirus Cap1_SP_115
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
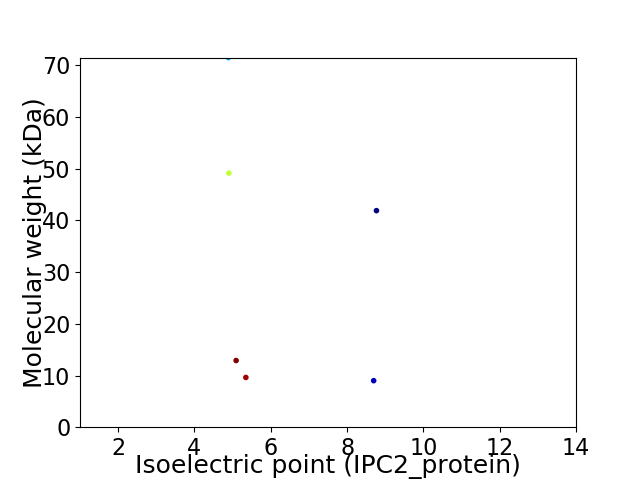
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
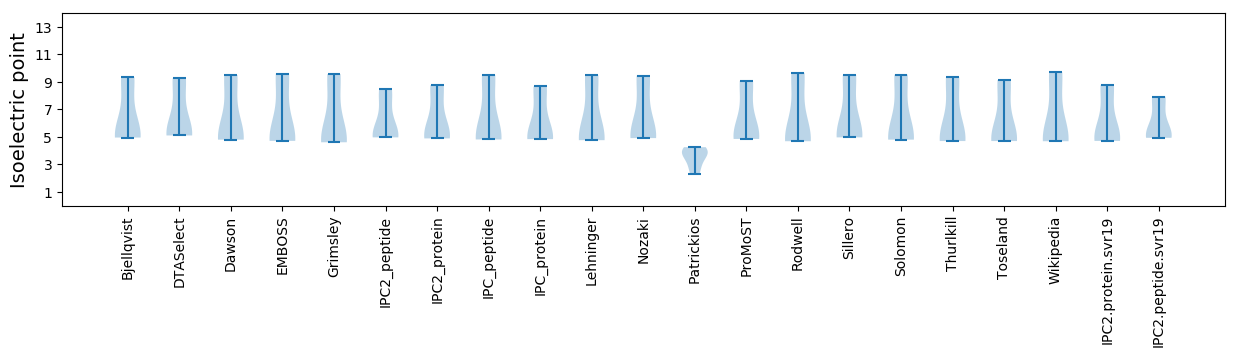
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4F8|A0A4P8W4F8_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_115 OX=2584808 PE=4 SV=1
MM1 pKa = 7.36ATNPLQPANHH11 pKa = 7.08AEE13 pKa = 4.21TLNPEE18 pKa = 4.06AQANNPSIRR27 pKa = 11.84FRR29 pKa = 11.84SPSNTFYY36 pKa = 10.96LPKK39 pKa = 9.99RR40 pKa = 11.84HH41 pKa = 6.39YY42 pKa = 9.21DD43 pKa = 3.49TIRR46 pKa = 11.84FGEE49 pKa = 4.01YY50 pKa = 9.48RR51 pKa = 11.84PVYY54 pKa = 8.82NLEE57 pKa = 4.26MIPNGRR63 pKa = 11.84YY64 pKa = 8.25MLQDD68 pKa = 3.32ANRR71 pKa = 11.84IEE73 pKa = 4.49SFTFASRR80 pKa = 11.84FMEE83 pKa = 4.27GLSYY87 pKa = 10.61TKK89 pKa = 10.36DD90 pKa = 3.36YY91 pKa = 10.96FQVPFSAILPRR102 pKa = 11.84NWEE105 pKa = 3.94KK106 pKa = 10.57IYY108 pKa = 10.63RR109 pKa = 11.84NPAFGDD115 pKa = 3.87DD116 pKa = 3.95VPDD119 pKa = 5.19DD120 pKa = 4.32SMSLINFQKK129 pKa = 10.49LFNDD133 pKa = 3.19IASRR137 pKa = 11.84LISQPSSLQDD147 pKa = 3.36LTFMLILPYY156 pKa = 10.46LFSRR160 pKa = 11.84GSLLDD165 pKa = 3.38SLGFPVVDD173 pKa = 5.14ISFLDD178 pKa = 3.78DD179 pKa = 3.49NGEE182 pKa = 3.77YY183 pKa = 10.5DD184 pKa = 4.43IIQEE188 pKa = 4.5IIDD191 pKa = 3.81TSGSFQLKK199 pKa = 10.3SPISSIPNFFYY210 pKa = 10.61KK211 pKa = 10.73VSDD214 pKa = 3.17ASTFNILINDD224 pKa = 4.39FLLNGYY230 pKa = 9.22SIISGPSDD238 pKa = 3.53FEE240 pKa = 4.27SLRR243 pKa = 11.84PLSAQWKK250 pKa = 9.91AKK252 pKa = 9.35FQALASSLAGSEE264 pKa = 4.53LISVASLYY272 pKa = 10.56AYY274 pKa = 9.95KK275 pKa = 9.37LTCAKK280 pKa = 10.39FYY282 pKa = 10.89TNDD285 pKa = 3.85NIDD288 pKa = 4.88PIYY291 pKa = 10.14MSQLWIDD298 pKa = 3.91SVQSLLSVSDD308 pKa = 3.54DD309 pKa = 3.61TQPTFNYY316 pKa = 10.13NGSKK320 pKa = 10.4YY321 pKa = 10.5LYY323 pKa = 10.12DD324 pKa = 3.9VLSLHH329 pKa = 6.74YY330 pKa = 10.38INEE333 pKa = 4.32KK334 pKa = 10.57LFSLLDD340 pKa = 3.56SDD342 pKa = 4.2YY343 pKa = 10.77TNALIALFGPEE354 pKa = 4.4HH355 pKa = 7.48VLIQQDD361 pKa = 3.93FFLSARR367 pKa = 11.84SRR369 pKa = 11.84PLAVGNVDD377 pKa = 2.82IHH379 pKa = 7.09VNTSSQSVSAIDD391 pKa = 3.37VTKK394 pKa = 10.89NITIQRR400 pKa = 11.84YY401 pKa = 8.23LNAVNRR407 pKa = 11.84LGNKK411 pKa = 6.29ITSYY415 pKa = 10.31MEE417 pKa = 5.29GIFSTQPQEE426 pKa = 4.27CPEE429 pKa = 4.19PVLLTSLRR437 pKa = 11.84YY438 pKa = 9.53DD439 pKa = 3.06VSTNQTTNTANDD451 pKa = 3.54QGNVTSLAQSNGSKK465 pKa = 10.23YY466 pKa = 10.9AFDD469 pKa = 4.08IFCSEE474 pKa = 3.94PSILLGIAHH483 pKa = 6.64FAIPKK488 pKa = 8.85VYY490 pKa = 10.08RR491 pKa = 11.84FAMDD495 pKa = 2.97MCKK498 pKa = 10.21YY499 pKa = 9.51HH500 pKa = 7.3ANRR503 pKa = 11.84FQFFNPMLEE512 pKa = 4.28NIGDD516 pKa = 3.83DD517 pKa = 4.82GIPVQCFDD525 pKa = 4.13FRR527 pKa = 11.84MPKK530 pKa = 10.3NLDD533 pKa = 3.45KK534 pKa = 11.36PNFGYY539 pKa = 8.42ITRR542 pKa = 11.84FGEE545 pKa = 4.13YY546 pKa = 10.45KK547 pKa = 9.87LTFDD551 pKa = 3.84EE552 pKa = 4.28AHH554 pKa = 6.56GGFMTNPLQSWIYY567 pKa = 10.37QGLDD571 pKa = 3.08ALDD574 pKa = 4.16AQSVINPFVIRR585 pKa = 11.84YY586 pKa = 8.5RR587 pKa = 11.84VGSMDD592 pKa = 3.65YY593 pKa = 10.24LYY595 pKa = 11.1SAVSPYY601 pKa = 9.93GPQWYY606 pKa = 8.93YY607 pKa = 11.22HH608 pKa = 6.08FIVRR612 pKa = 11.84NEE614 pKa = 4.11TNLTANLPLSYY625 pKa = 11.0NPGILL630 pKa = 3.62
MM1 pKa = 7.36ATNPLQPANHH11 pKa = 7.08AEE13 pKa = 4.21TLNPEE18 pKa = 4.06AQANNPSIRR27 pKa = 11.84FRR29 pKa = 11.84SPSNTFYY36 pKa = 10.96LPKK39 pKa = 9.99RR40 pKa = 11.84HH41 pKa = 6.39YY42 pKa = 9.21DD43 pKa = 3.49TIRR46 pKa = 11.84FGEE49 pKa = 4.01YY50 pKa = 9.48RR51 pKa = 11.84PVYY54 pKa = 8.82NLEE57 pKa = 4.26MIPNGRR63 pKa = 11.84YY64 pKa = 8.25MLQDD68 pKa = 3.32ANRR71 pKa = 11.84IEE73 pKa = 4.49SFTFASRR80 pKa = 11.84FMEE83 pKa = 4.27GLSYY87 pKa = 10.61TKK89 pKa = 10.36DD90 pKa = 3.36YY91 pKa = 10.96FQVPFSAILPRR102 pKa = 11.84NWEE105 pKa = 3.94KK106 pKa = 10.57IYY108 pKa = 10.63RR109 pKa = 11.84NPAFGDD115 pKa = 3.87DD116 pKa = 3.95VPDD119 pKa = 5.19DD120 pKa = 4.32SMSLINFQKK129 pKa = 10.49LFNDD133 pKa = 3.19IASRR137 pKa = 11.84LISQPSSLQDD147 pKa = 3.36LTFMLILPYY156 pKa = 10.46LFSRR160 pKa = 11.84GSLLDD165 pKa = 3.38SLGFPVVDD173 pKa = 5.14ISFLDD178 pKa = 3.78DD179 pKa = 3.49NGEE182 pKa = 3.77YY183 pKa = 10.5DD184 pKa = 4.43IIQEE188 pKa = 4.5IIDD191 pKa = 3.81TSGSFQLKK199 pKa = 10.3SPISSIPNFFYY210 pKa = 10.61KK211 pKa = 10.73VSDD214 pKa = 3.17ASTFNILINDD224 pKa = 4.39FLLNGYY230 pKa = 9.22SIISGPSDD238 pKa = 3.53FEE240 pKa = 4.27SLRR243 pKa = 11.84PLSAQWKK250 pKa = 9.91AKK252 pKa = 9.35FQALASSLAGSEE264 pKa = 4.53LISVASLYY272 pKa = 10.56AYY274 pKa = 9.95KK275 pKa = 9.37LTCAKK280 pKa = 10.39FYY282 pKa = 10.89TNDD285 pKa = 3.85NIDD288 pKa = 4.88PIYY291 pKa = 10.14MSQLWIDD298 pKa = 3.91SVQSLLSVSDD308 pKa = 3.54DD309 pKa = 3.61TQPTFNYY316 pKa = 10.13NGSKK320 pKa = 10.4YY321 pKa = 10.5LYY323 pKa = 10.12DD324 pKa = 3.9VLSLHH329 pKa = 6.74YY330 pKa = 10.38INEE333 pKa = 4.32KK334 pKa = 10.57LFSLLDD340 pKa = 3.56SDD342 pKa = 4.2YY343 pKa = 10.77TNALIALFGPEE354 pKa = 4.4HH355 pKa = 7.48VLIQQDD361 pKa = 3.93FFLSARR367 pKa = 11.84SRR369 pKa = 11.84PLAVGNVDD377 pKa = 2.82IHH379 pKa = 7.09VNTSSQSVSAIDD391 pKa = 3.37VTKK394 pKa = 10.89NITIQRR400 pKa = 11.84YY401 pKa = 8.23LNAVNRR407 pKa = 11.84LGNKK411 pKa = 6.29ITSYY415 pKa = 10.31MEE417 pKa = 5.29GIFSTQPQEE426 pKa = 4.27CPEE429 pKa = 4.19PVLLTSLRR437 pKa = 11.84YY438 pKa = 9.53DD439 pKa = 3.06VSTNQTTNTANDD451 pKa = 3.54QGNVTSLAQSNGSKK465 pKa = 10.23YY466 pKa = 10.9AFDD469 pKa = 4.08IFCSEE474 pKa = 3.94PSILLGIAHH483 pKa = 6.64FAIPKK488 pKa = 8.85VYY490 pKa = 10.08RR491 pKa = 11.84FAMDD495 pKa = 2.97MCKK498 pKa = 10.21YY499 pKa = 9.51HH500 pKa = 7.3ANRR503 pKa = 11.84FQFFNPMLEE512 pKa = 4.28NIGDD516 pKa = 3.83DD517 pKa = 4.82GIPVQCFDD525 pKa = 4.13FRR527 pKa = 11.84MPKK530 pKa = 10.3NLDD533 pKa = 3.45KK534 pKa = 11.36PNFGYY539 pKa = 8.42ITRR542 pKa = 11.84FGEE545 pKa = 4.13YY546 pKa = 10.45KK547 pKa = 9.87LTFDD551 pKa = 3.84EE552 pKa = 4.28AHH554 pKa = 6.56GGFMTNPLQSWIYY567 pKa = 10.37QGLDD571 pKa = 3.08ALDD574 pKa = 4.16AQSVINPFVIRR585 pKa = 11.84YY586 pKa = 8.5RR587 pKa = 11.84VGSMDD592 pKa = 3.65YY593 pKa = 10.24LYY595 pKa = 11.1SAVSPYY601 pKa = 9.93GPQWYY606 pKa = 8.93YY607 pKa = 11.22HH608 pKa = 6.08FIVRR612 pKa = 11.84NEE614 pKa = 4.11TNLTANLPLSYY625 pKa = 11.0NPGILL630 pKa = 3.62
Molecular weight: 71.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7U3|A0A4P8W7U3_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_115 OX=2584808 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 9.93KK3 pKa = 10.08VCVFDD8 pKa = 4.83CPPGMYY14 pKa = 10.27YY15 pKa = 10.32EE16 pKa = 4.47WLPHH20 pKa = 5.74SCIKK24 pKa = 10.42YY25 pKa = 8.78VRR27 pKa = 11.84KK28 pKa = 9.97VRR30 pKa = 11.84NNSNKK35 pKa = 8.94FHH37 pKa = 7.6AIRR40 pKa = 11.84YY41 pKa = 5.74TRR43 pKa = 11.84WLDD46 pKa = 3.25GLIYY50 pKa = 9.8TEE52 pKa = 4.21KK53 pKa = 9.78TVCSLCFLVPLGVNYY68 pKa = 10.46NPYY71 pKa = 10.03KK72 pKa = 10.46QLFF75 pKa = 3.65
MM1 pKa = 7.48KK2 pKa = 9.93KK3 pKa = 10.08VCVFDD8 pKa = 4.83CPPGMYY14 pKa = 10.27YY15 pKa = 10.32EE16 pKa = 4.47WLPHH20 pKa = 5.74SCIKK24 pKa = 10.42YY25 pKa = 8.78VRR27 pKa = 11.84KK28 pKa = 9.97VRR30 pKa = 11.84NNSNKK35 pKa = 8.94FHH37 pKa = 7.6AIRR40 pKa = 11.84YY41 pKa = 5.74TRR43 pKa = 11.84WLDD46 pKa = 3.25GLIYY50 pKa = 9.8TEE52 pKa = 4.21KK53 pKa = 9.78TVCSLCFLVPLGVNYY68 pKa = 10.46NPYY71 pKa = 10.03KK72 pKa = 10.46QLFF75 pKa = 3.65
Molecular weight: 9.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1702 |
75 |
630 |
283.7 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.463 ± 1.747 | 1.234 ± 0.655 |
6.169 ± 0.404 | 3.995 ± 0.477 |
6.639 ± 0.501 | 4.407 ± 0.406 |
1.528 ± 0.319 | 6.169 ± 0.592 |
5.464 ± 1.042 | 9.166 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.181 | 7.873 ± 0.384 |
4.759 ± 0.717 | 4.407 ± 1.058 |
4.054 ± 0.501 | 10.106 ± 1.064 |
4.113 ± 0.372 | 4.7 ± 0.47 |
0.881 ± 0.233 | 6.052 ± 0.406 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
