
Sphingobacterium sp. ML3W
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Sphingobacterium; unclassified Sphingobacterium
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
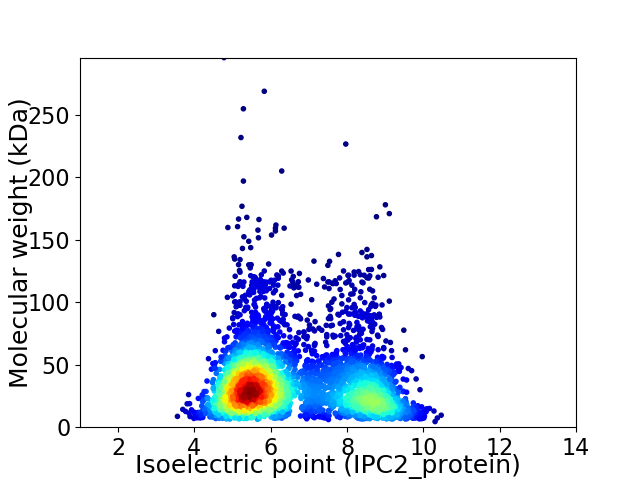
Virtual 2D-PAGE plot for 4058 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A088EZZ5|A0A088EZZ5_9SPHI Thymidylate synthase OS=Sphingobacterium sp. ML3W OX=1538644 GN=KO02_16490 PE=4 SV=1
MM1 pKa = 7.56VIPNSLADD9 pKa = 5.25AEE11 pKa = 5.03LLLNDD16 pKa = 4.21YY17 pKa = 10.23STMNTGYY24 pKa = 9.4PVYY27 pKa = 10.79GEE29 pKa = 4.66LSADD33 pKa = 3.54EE34 pKa = 4.86YY35 pKa = 11.6YY36 pKa = 11.08VALEE40 pKa = 4.43TFDD43 pKa = 4.6GMLDD47 pKa = 3.77FDD49 pKa = 4.94QRR51 pKa = 11.84NTYY54 pKa = 7.71TWMDD58 pKa = 3.92IIYY61 pKa = 10.29DD62 pKa = 5.43DD63 pKa = 3.64VAQWQRR69 pKa = 11.84PYY71 pKa = 10.97KK72 pKa = 10.62AVFNANQALEE82 pKa = 4.4IINNNSADD90 pKa = 3.59TKK92 pKa = 10.87IDD94 pKa = 3.12IKK96 pKa = 10.96KK97 pKa = 10.48
MM1 pKa = 7.56VIPNSLADD9 pKa = 5.25AEE11 pKa = 5.03LLLNDD16 pKa = 4.21YY17 pKa = 10.23STMNTGYY24 pKa = 9.4PVYY27 pKa = 10.79GEE29 pKa = 4.66LSADD33 pKa = 3.54EE34 pKa = 4.86YY35 pKa = 11.6YY36 pKa = 11.08VALEE40 pKa = 4.43TFDD43 pKa = 4.6GMLDD47 pKa = 3.77FDD49 pKa = 4.94QRR51 pKa = 11.84NTYY54 pKa = 7.71TWMDD58 pKa = 3.92IIYY61 pKa = 10.29DD62 pKa = 5.43DD63 pKa = 3.64VAQWQRR69 pKa = 11.84PYY71 pKa = 10.97KK72 pKa = 10.62AVFNANQALEE82 pKa = 4.4IINNNSADD90 pKa = 3.59TKK92 pKa = 10.87IDD94 pKa = 3.12IKK96 pKa = 10.96KK97 pKa = 10.48
Molecular weight: 11.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088EUE5|A0A088EUE5_9SPHI Uncharacterized protein OS=Sphingobacterium sp. ML3W OX=1538644 GN=KO02_04745 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.14RR3 pKa = 11.84IKK5 pKa = 10.03HH6 pKa = 5.7IKK8 pKa = 9.63LIAIAIFSILFFANPNTTFAQRR30 pKa = 11.84GYY32 pKa = 11.06NDD34 pKa = 3.57YY35 pKa = 11.06QSGVSFQMFYY45 pKa = 11.45DD46 pKa = 3.88EE47 pKa = 4.67LAPYY51 pKa = 9.74GDD53 pKa = 3.28WVNDD57 pKa = 3.78RR58 pKa = 11.84EE59 pKa = 4.53HH60 pKa = 8.17GYY62 pKa = 8.18MWIPDD67 pKa = 3.92VGPNFQPYY75 pKa = 8.14STNGYY80 pKa = 6.2WTMTEE85 pKa = 4.29YY86 pKa = 11.42GNTWVSNYY94 pKa = 8.74SWGWAPFHH102 pKa = 6.16YY103 pKa = 10.38GRR105 pKa = 11.84WEE107 pKa = 3.81YY108 pKa = 10.92NNNYY112 pKa = 7.86GWAWTPDD119 pKa = 4.17YY120 pKa = 10.5EE121 pKa = 5.28WGPAWVNWRR130 pKa = 11.84QGSGYY135 pKa = 10.24YY136 pKa = 10.3GWAPLGINISINLPMNLWVFVGSSNIYY163 pKa = 9.4SNRR166 pKa = 11.84LDD168 pKa = 3.17RR169 pKa = 11.84YY170 pKa = 8.25YY171 pKa = 11.23VHH173 pKa = 7.11PRR175 pKa = 11.84NYY177 pKa = 10.47NNFYY181 pKa = 11.13NKK183 pKa = 7.88TTIINNTIIVNNRR196 pKa = 11.84NYY198 pKa = 10.46YY199 pKa = 9.53GGPRR203 pKa = 11.84RR204 pKa = 11.84SDD206 pKa = 3.05IEE208 pKa = 3.95RR209 pKa = 11.84STGKK213 pKa = 10.05RR214 pKa = 11.84VSVRR218 pKa = 11.84NINNSGRR225 pKa = 11.84AGTSRR230 pKa = 11.84VTKK233 pKa = 10.67NSIDD237 pKa = 3.52MYY239 pKa = 10.68RR240 pKa = 11.84PNIDD244 pKa = 3.24RR245 pKa = 11.84NSRR248 pKa = 11.84TDD250 pKa = 3.19ARR252 pKa = 11.84PSRR255 pKa = 11.84IAEE258 pKa = 4.02ASSRR262 pKa = 11.84TRR264 pKa = 11.84NNNTSLQNRR273 pKa = 11.84NDD275 pKa = 3.43KK276 pKa = 10.92VISDD280 pKa = 4.25RR281 pKa = 11.84DD282 pKa = 3.62NMTSTRR288 pKa = 11.84GNRR291 pKa = 11.84EE292 pKa = 3.65LYY294 pKa = 10.06IDD296 pKa = 3.65NKK298 pKa = 10.83GNATVRR304 pKa = 11.84SGSNNSTTTRR314 pKa = 11.84NNDD317 pKa = 2.68RR318 pKa = 11.84AANRR322 pKa = 11.84NQNSSNVTGSSRR334 pKa = 11.84TRR336 pKa = 11.84DD337 pKa = 3.35NSNATVNNSSTTGNARR353 pKa = 11.84TEE355 pKa = 4.49TNSSSVRR362 pKa = 11.84TDD364 pKa = 2.91RR365 pKa = 11.84TRR367 pKa = 11.84GNTSSSKK374 pKa = 10.04QEE376 pKa = 4.16EE377 pKa = 4.34IRR379 pKa = 11.84KK380 pKa = 9.71SGQPVQGQATRR391 pKa = 11.84SSRR394 pKa = 11.84TTQTQPARR402 pKa = 11.84NEE404 pKa = 3.79QPAKK408 pKa = 10.35VQSTSNDD415 pKa = 2.69ARR417 pKa = 11.84TSRR420 pKa = 11.84PAQGQQSRR428 pKa = 11.84SAQPAKK434 pKa = 10.46VQSSTSNDD442 pKa = 2.72ARR444 pKa = 11.84TSRR447 pKa = 11.84PAQGQQTRR455 pKa = 11.84SAQPARR461 pKa = 11.84VQSSTTDD468 pKa = 3.04RR469 pKa = 11.84GSSSSRR475 pKa = 11.84SSSSTVEE482 pKa = 4.03RR483 pKa = 11.84NTSQSEE489 pKa = 4.19NNRR492 pKa = 11.84SSGSRR497 pKa = 11.84TRR499 pKa = 3.72
MM1 pKa = 7.43KK2 pKa = 10.14RR3 pKa = 11.84IKK5 pKa = 10.03HH6 pKa = 5.7IKK8 pKa = 9.63LIAIAIFSILFFANPNTTFAQRR30 pKa = 11.84GYY32 pKa = 11.06NDD34 pKa = 3.57YY35 pKa = 11.06QSGVSFQMFYY45 pKa = 11.45DD46 pKa = 3.88EE47 pKa = 4.67LAPYY51 pKa = 9.74GDD53 pKa = 3.28WVNDD57 pKa = 3.78RR58 pKa = 11.84EE59 pKa = 4.53HH60 pKa = 8.17GYY62 pKa = 8.18MWIPDD67 pKa = 3.92VGPNFQPYY75 pKa = 8.14STNGYY80 pKa = 6.2WTMTEE85 pKa = 4.29YY86 pKa = 11.42GNTWVSNYY94 pKa = 8.74SWGWAPFHH102 pKa = 6.16YY103 pKa = 10.38GRR105 pKa = 11.84WEE107 pKa = 3.81YY108 pKa = 10.92NNNYY112 pKa = 7.86GWAWTPDD119 pKa = 4.17YY120 pKa = 10.5EE121 pKa = 5.28WGPAWVNWRR130 pKa = 11.84QGSGYY135 pKa = 10.24YY136 pKa = 10.3GWAPLGINISINLPMNLWVFVGSSNIYY163 pKa = 9.4SNRR166 pKa = 11.84LDD168 pKa = 3.17RR169 pKa = 11.84YY170 pKa = 8.25YY171 pKa = 11.23VHH173 pKa = 7.11PRR175 pKa = 11.84NYY177 pKa = 10.47NNFYY181 pKa = 11.13NKK183 pKa = 7.88TTIINNTIIVNNRR196 pKa = 11.84NYY198 pKa = 10.46YY199 pKa = 9.53GGPRR203 pKa = 11.84RR204 pKa = 11.84SDD206 pKa = 3.05IEE208 pKa = 3.95RR209 pKa = 11.84STGKK213 pKa = 10.05RR214 pKa = 11.84VSVRR218 pKa = 11.84NINNSGRR225 pKa = 11.84AGTSRR230 pKa = 11.84VTKK233 pKa = 10.67NSIDD237 pKa = 3.52MYY239 pKa = 10.68RR240 pKa = 11.84PNIDD244 pKa = 3.24RR245 pKa = 11.84NSRR248 pKa = 11.84TDD250 pKa = 3.19ARR252 pKa = 11.84PSRR255 pKa = 11.84IAEE258 pKa = 4.02ASSRR262 pKa = 11.84TRR264 pKa = 11.84NNNTSLQNRR273 pKa = 11.84NDD275 pKa = 3.43KK276 pKa = 10.92VISDD280 pKa = 4.25RR281 pKa = 11.84DD282 pKa = 3.62NMTSTRR288 pKa = 11.84GNRR291 pKa = 11.84EE292 pKa = 3.65LYY294 pKa = 10.06IDD296 pKa = 3.65NKK298 pKa = 10.83GNATVRR304 pKa = 11.84SGSNNSTTTRR314 pKa = 11.84NNDD317 pKa = 2.68RR318 pKa = 11.84AANRR322 pKa = 11.84NQNSSNVTGSSRR334 pKa = 11.84TRR336 pKa = 11.84DD337 pKa = 3.35NSNATVNNSSTTGNARR353 pKa = 11.84TEE355 pKa = 4.49TNSSSVRR362 pKa = 11.84TDD364 pKa = 2.91RR365 pKa = 11.84TRR367 pKa = 11.84GNTSSSKK374 pKa = 10.04QEE376 pKa = 4.16EE377 pKa = 4.34IRR379 pKa = 11.84KK380 pKa = 9.71SGQPVQGQATRR391 pKa = 11.84SSRR394 pKa = 11.84TTQTQPARR402 pKa = 11.84NEE404 pKa = 3.79QPAKK408 pKa = 10.35VQSTSNDD415 pKa = 2.69ARR417 pKa = 11.84TSRR420 pKa = 11.84PAQGQQSRR428 pKa = 11.84SAQPAKK434 pKa = 10.46VQSSTSNDD442 pKa = 2.72ARR444 pKa = 11.84TSRR447 pKa = 11.84PAQGQQTRR455 pKa = 11.84SAQPARR461 pKa = 11.84VQSSTTDD468 pKa = 3.04RR469 pKa = 11.84GSSSSRR475 pKa = 11.84SSSSTVEE482 pKa = 4.03RR483 pKa = 11.84NTSQSEE489 pKa = 4.19NNRR492 pKa = 11.84SSGSRR497 pKa = 11.84TRR499 pKa = 3.72
Molecular weight: 56.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1400076 |
38 |
2700 |
345.0 |
38.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.857 ± 0.041 | 0.731 ± 0.011 |
5.566 ± 0.028 | 6.093 ± 0.038 |
4.95 ± 0.027 | 6.529 ± 0.039 |
1.9 ± 0.015 | 7.73 ± 0.039 |
7.242 ± 0.035 | 9.57 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.336 ± 0.018 | 5.75 ± 0.034 |
3.492 ± 0.021 | 3.88 ± 0.025 |
3.831 ± 0.023 | 6.536 ± 0.031 |
5.485 ± 0.024 | 6.16 ± 0.026 |
1.166 ± 0.015 | 4.197 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
