
Garba virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sunrhavirus; Garba sunrhavirus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
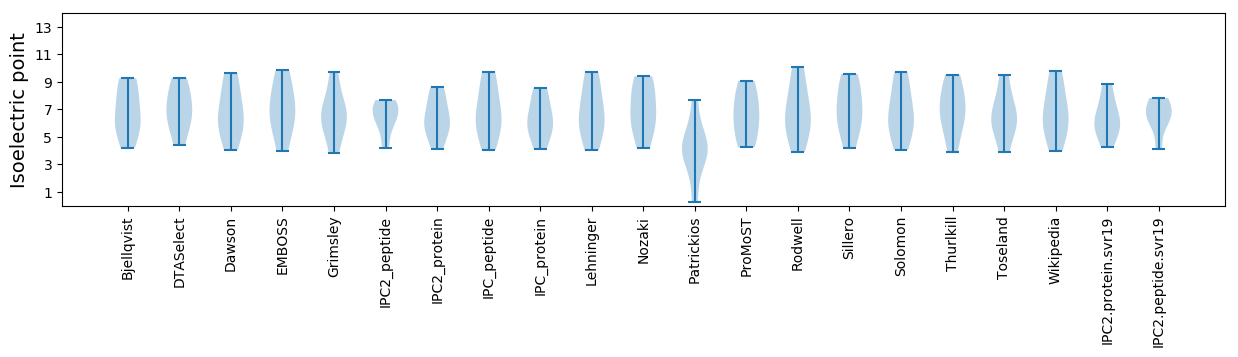
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R119|A0A0D3R119_9RHAB Glycoprotein OS=Garba virus OX=864696 PE=4 SV=1
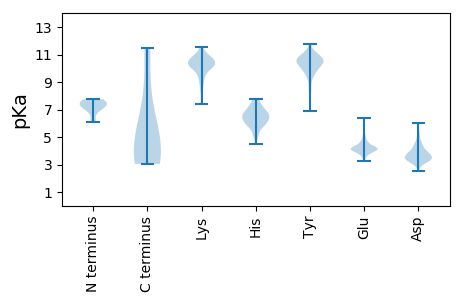
MM1 pKa = 7.29NNHH4 pKa = 6.29RR5 pKa = 11.84AKK7 pKa = 10.25EE8 pKa = 3.8IEE10 pKa = 4.4KK11 pKa = 10.17IYY13 pKa = 10.94EE14 pKa = 3.93NLKK17 pKa = 10.53NEE19 pKa = 4.14PSLCGDD25 pKa = 3.82DD26 pKa = 5.38SEE28 pKa = 5.31DD29 pKa = 3.39TGGMYY34 pKa = 10.92GLGMPGTSTGSPLLQSIQMGEE55 pKa = 4.06VHH57 pKa = 7.64DD58 pKa = 4.49SRR60 pKa = 11.84TTSEE64 pKa = 4.14SQKK67 pKa = 10.01PVPWDD72 pKa = 3.28SDD74 pKa = 3.48EE75 pKa = 6.37DD76 pKa = 4.27SDD78 pKa = 4.64TDD80 pKa = 3.56FVYY83 pKa = 10.31PSRR86 pKa = 11.84DD87 pKa = 3.42KK88 pKa = 11.49GCSSDD93 pKa = 5.2DD94 pKa = 3.7EE95 pKa = 4.69NSDD98 pKa = 3.99GEE100 pKa = 4.63TTKK103 pKa = 10.71PSADD107 pKa = 3.67FARR110 pKa = 11.84GGYY113 pKa = 10.51GDD115 pKa = 4.14GFRR118 pKa = 11.84DD119 pKa = 3.71GMRR122 pKa = 11.84EE123 pKa = 3.86AVSKK127 pKa = 10.74INYY130 pKa = 9.43LLTTQGVGLFHH141 pKa = 7.58FDD143 pKa = 3.47GADD146 pKa = 2.93IFIKK150 pKa = 10.69KK151 pKa = 7.37EE152 pKa = 3.73TSLNIEE158 pKa = 3.95NQVDD162 pKa = 4.72GEE164 pKa = 4.5CSLTTCQTDD173 pKa = 3.13QRR175 pKa = 11.84KK176 pKa = 10.1DD177 pKa = 3.17IEE179 pKa = 4.22PRR181 pKa = 11.84EE182 pKa = 4.15MKK184 pKa = 10.0QDD186 pKa = 3.61SASTEE191 pKa = 4.04MDD193 pKa = 3.4SDD195 pKa = 3.69TDD197 pKa = 3.78DD198 pKa = 4.82PEE200 pKa = 5.05LVSSDD205 pKa = 3.67EE206 pKa = 4.0FLEE209 pKa = 4.12ILDD212 pKa = 4.53GGLCVEE218 pKa = 5.36LGGDD222 pKa = 4.11SKK224 pKa = 11.06FLSLDD229 pKa = 3.24SVNGVKK235 pKa = 10.4FEE237 pKa = 4.4EE238 pKa = 4.35NKK240 pKa = 9.53NSKK243 pKa = 7.87MTVVDD248 pKa = 4.32WVLKK252 pKa = 10.63LCTT255 pKa = 4.01
MM1 pKa = 7.29NNHH4 pKa = 6.29RR5 pKa = 11.84AKK7 pKa = 10.25EE8 pKa = 3.8IEE10 pKa = 4.4KK11 pKa = 10.17IYY13 pKa = 10.94EE14 pKa = 3.93NLKK17 pKa = 10.53NEE19 pKa = 4.14PSLCGDD25 pKa = 3.82DD26 pKa = 5.38SEE28 pKa = 5.31DD29 pKa = 3.39TGGMYY34 pKa = 10.92GLGMPGTSTGSPLLQSIQMGEE55 pKa = 4.06VHH57 pKa = 7.64DD58 pKa = 4.49SRR60 pKa = 11.84TTSEE64 pKa = 4.14SQKK67 pKa = 10.01PVPWDD72 pKa = 3.28SDD74 pKa = 3.48EE75 pKa = 6.37DD76 pKa = 4.27SDD78 pKa = 4.64TDD80 pKa = 3.56FVYY83 pKa = 10.31PSRR86 pKa = 11.84DD87 pKa = 3.42KK88 pKa = 11.49GCSSDD93 pKa = 5.2DD94 pKa = 3.7EE95 pKa = 4.69NSDD98 pKa = 3.99GEE100 pKa = 4.63TTKK103 pKa = 10.71PSADD107 pKa = 3.67FARR110 pKa = 11.84GGYY113 pKa = 10.51GDD115 pKa = 4.14GFRR118 pKa = 11.84DD119 pKa = 3.71GMRR122 pKa = 11.84EE123 pKa = 3.86AVSKK127 pKa = 10.74INYY130 pKa = 9.43LLTTQGVGLFHH141 pKa = 7.58FDD143 pKa = 3.47GADD146 pKa = 2.93IFIKK150 pKa = 10.69KK151 pKa = 7.37EE152 pKa = 3.73TSLNIEE158 pKa = 3.95NQVDD162 pKa = 4.72GEE164 pKa = 4.5CSLTTCQTDD173 pKa = 3.13QRR175 pKa = 11.84KK176 pKa = 10.1DD177 pKa = 3.17IEE179 pKa = 4.22PRR181 pKa = 11.84EE182 pKa = 4.15MKK184 pKa = 10.0QDD186 pKa = 3.61SASTEE191 pKa = 4.04MDD193 pKa = 3.4SDD195 pKa = 3.69TDD197 pKa = 3.78DD198 pKa = 4.82PEE200 pKa = 5.05LVSSDD205 pKa = 3.67EE206 pKa = 4.0FLEE209 pKa = 4.12ILDD212 pKa = 4.53GGLCVEE218 pKa = 5.36LGGDD222 pKa = 4.11SKK224 pKa = 11.06FLSLDD229 pKa = 3.24SVNGVKK235 pKa = 10.4FEE237 pKa = 4.4EE238 pKa = 4.35NKK240 pKa = 9.53NSKK243 pKa = 7.87MTVVDD248 pKa = 4.32WVLKK252 pKa = 10.63LCTT255 pKa = 4.01
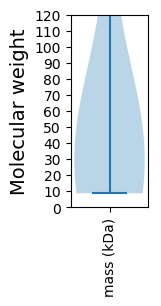
Molecular weight: 28.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R0Y6|A0A0D3R0Y6_9RHAB Phosphoprotein OS=Garba virus OX=864696 PE=4 SV=1
MM1 pKa = 7.2NRR3 pKa = 11.84LKK5 pKa = 11.01RR6 pKa = 11.84MFGYY10 pKa = 10.71GEE12 pKa = 4.11EE13 pKa = 4.27DD14 pKa = 3.15LADD17 pKa = 3.84FVGIEE22 pKa = 4.21SKK24 pKa = 10.09MVTVVEE30 pKa = 4.27VKK32 pKa = 10.19IQFSLNVTISRR43 pKa = 11.84VGKK46 pKa = 9.45SALSRR51 pKa = 11.84EE52 pKa = 4.4IILKK56 pKa = 10.01EE57 pKa = 3.63LLSNYY62 pKa = 9.58RR63 pKa = 11.84GPSEE67 pKa = 4.35KK68 pKa = 10.19EE69 pKa = 3.49ALFVIGSLLSVPSWIRR85 pKa = 11.84KK86 pKa = 8.99KK87 pKa = 10.76DD88 pKa = 3.3GSGYY92 pKa = 9.75GFRR95 pKa = 11.84GEE97 pKa = 4.11FGIKK101 pKa = 10.21FEE103 pKa = 4.48TDD105 pKa = 3.02NLSLSNNFEE114 pKa = 4.23HH115 pKa = 7.17EE116 pKa = 4.55SISTYY121 pKa = 11.05VKK123 pKa = 10.68DD124 pKa = 3.27NWIVIINSSVKK135 pKa = 8.93TKK137 pKa = 10.33RR138 pKa = 11.84GKK140 pKa = 10.84GGFNPATKK148 pKa = 10.17LQEE151 pKa = 3.96ILSKK155 pKa = 10.98FEE157 pKa = 3.52IRR159 pKa = 11.84TNLFDD164 pKa = 4.37FSPIFIVHH172 pKa = 6.68
MM1 pKa = 7.2NRR3 pKa = 11.84LKK5 pKa = 11.01RR6 pKa = 11.84MFGYY10 pKa = 10.71GEE12 pKa = 4.11EE13 pKa = 4.27DD14 pKa = 3.15LADD17 pKa = 3.84FVGIEE22 pKa = 4.21SKK24 pKa = 10.09MVTVVEE30 pKa = 4.27VKK32 pKa = 10.19IQFSLNVTISRR43 pKa = 11.84VGKK46 pKa = 9.45SALSRR51 pKa = 11.84EE52 pKa = 4.4IILKK56 pKa = 10.01EE57 pKa = 3.63LLSNYY62 pKa = 9.58RR63 pKa = 11.84GPSEE67 pKa = 4.35KK68 pKa = 10.19EE69 pKa = 3.49ALFVIGSLLSVPSWIRR85 pKa = 11.84KK86 pKa = 8.99KK87 pKa = 10.76DD88 pKa = 3.3GSGYY92 pKa = 9.75GFRR95 pKa = 11.84GEE97 pKa = 4.11FGIKK101 pKa = 10.21FEE103 pKa = 4.48TDD105 pKa = 3.02NLSLSNNFEE114 pKa = 4.23HH115 pKa = 7.17EE116 pKa = 4.55SISTYY121 pKa = 11.05VKK123 pKa = 10.68DD124 pKa = 3.27NWIVIINSSVKK135 pKa = 8.93TKK137 pKa = 10.33RR138 pKa = 11.84GKK140 pKa = 10.84GGFNPATKK148 pKa = 10.17LQEE151 pKa = 3.96ILSKK155 pKa = 10.98FEE157 pKa = 3.52IRR159 pKa = 11.84TNLFDD164 pKa = 4.37FSPIFIVHH172 pKa = 6.68
Molecular weight: 19.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3663 |
71 |
2072 |
523.3 |
59.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.331 ± 0.781 | 2.075 ± 0.297 |
6.115 ± 0.82 | 6.416 ± 0.62 |
5.051 ± 0.366 | 5.842 ± 0.611 |
2.239 ± 0.372 | 7.289 ± 0.776 |
7.18 ± 0.276 | 9.801 ± 0.659 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.457 ± 0.247 | 6.115 ± 0.623 |
3.74 ± 0.225 | 3.221 ± 0.374 |
4.614 ± 0.441 | 7.835 ± 0.508 |
5.87 ± 0.389 | 5.597 ± 0.742 |
1.611 ± 0.138 | 3.604 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
