
Cellulomonas sp. SLBN-39
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; unclassified Cellulomonas
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
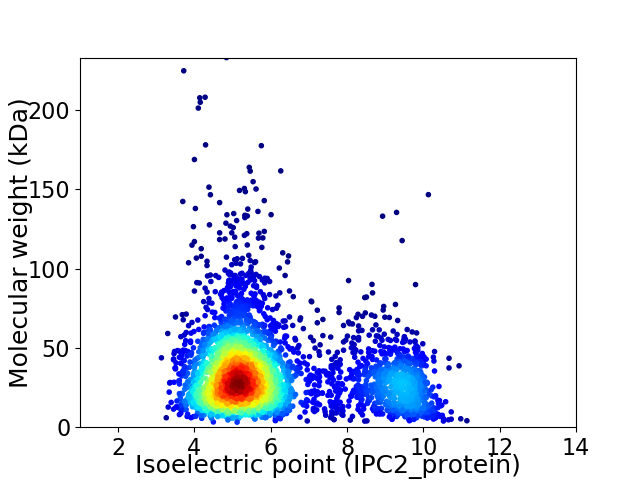
Virtual 2D-PAGE plot for 3709 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A542UX29|A0A542UX29_9CELL N-acetylneuraminate synthase OS=Cellulomonas sp. SLBN-39 OX=2768446 GN=FBY24_2753 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84MLAFAAAAAVLLTAACGGGDD24 pKa = 3.43EE25 pKa = 4.61AATGGEE31 pKa = 4.57TTGSTASLVLGQTDD45 pKa = 3.45IPASFDD51 pKa = 3.34VLGYY55 pKa = 11.16ASGTKK60 pKa = 7.66TQYY63 pKa = 8.72FTAVYY68 pKa = 7.78DD69 pKa = 3.62TLLRR73 pKa = 11.84QDD75 pKa = 3.17GSGAIVPGLATDD87 pKa = 3.82WAYY90 pKa = 11.21DD91 pKa = 3.53DD92 pKa = 3.87TRR94 pKa = 11.84TVLTLTLRR102 pKa = 11.84EE103 pKa = 4.29GVTFTDD109 pKa = 5.54GVALDD114 pKa = 4.14ADD116 pKa = 3.52AVVANIEE123 pKa = 4.18HH124 pKa = 6.36FRR126 pKa = 11.84GSSTPDD132 pKa = 3.32LSNAAYY138 pKa = 9.39IDD140 pKa = 3.92TVSAVDD146 pKa = 3.36ATTVEE151 pKa = 4.21ITLTEE156 pKa = 4.83PDD158 pKa = 3.86PMMLTWLTQPLGYY171 pKa = 9.47MSSPEE176 pKa = 3.7SWEE179 pKa = 4.17NEE181 pKa = 3.84DD182 pKa = 4.01VATNPVGSGPYY193 pKa = 10.27VLDD196 pKa = 3.51TEE198 pKa = 4.46ATVVGSTYY206 pKa = 10.93VFTKK210 pKa = 10.82NPDD213 pKa = 3.09YY214 pKa = 10.8WDD216 pKa = 5.22DD217 pKa = 3.71SLQLFDD223 pKa = 4.51TLTMNYY229 pKa = 9.97YY230 pKa = 10.83ADD232 pKa = 3.99QSALLNALQGGQVDD246 pKa = 4.11AAPFNQFSALEE257 pKa = 3.99QVEE260 pKa = 4.22AAGYY264 pKa = 7.96EE265 pKa = 4.66SNVSRR270 pKa = 11.84LDD272 pKa = 3.32WEE274 pKa = 4.63GLMLLDD280 pKa = 5.23RR281 pKa = 11.84DD282 pKa = 4.19GALDD286 pKa = 3.69PALADD291 pKa = 3.24VRR293 pKa = 11.84VRR295 pKa = 11.84QAINHH300 pKa = 7.16AIAKK304 pKa = 10.06DD305 pKa = 3.99SILSAILLDD314 pKa = 4.17HH315 pKa = 6.48GTATSQIFGPSTDD328 pKa = 4.53GYY330 pKa = 11.33DD331 pKa = 3.43EE332 pKa = 5.13SLDD335 pKa = 3.51GYY337 pKa = 8.47YY338 pKa = 10.2TYY340 pKa = 11.35DD341 pKa = 3.61PDD343 pKa = 4.12KK344 pKa = 11.4AKK346 pKa = 10.99ALLAEE351 pKa = 4.66AGYY354 pKa = 11.36ADD356 pKa = 4.14GLEE359 pKa = 4.14LTMPSTSVIDD369 pKa = 3.76QALLTSIQQQLADD382 pKa = 3.48VGITVSYY389 pKa = 10.34TDD391 pKa = 3.15VGTNFISDD399 pKa = 3.86LMGAKK404 pKa = 9.97YY405 pKa = 10.3GSSWMRR411 pKa = 11.84LASSSDD417 pKa = 2.77WQTATFALTPDD428 pKa = 3.25ALFNSFRR435 pKa = 11.84TQRR438 pKa = 11.84PEE440 pKa = 3.25VDD442 pKa = 3.46ALLEE446 pKa = 4.2TVQKK450 pKa = 9.77GTEE453 pKa = 3.98EE454 pKa = 3.91EE455 pKa = 3.88AAAAAQEE462 pKa = 4.06LNRR465 pKa = 11.84YY466 pKa = 7.53VVEE469 pKa = 4.35EE470 pKa = 3.78AWFAPFYY477 pKa = 10.73FVDD480 pKa = 3.37NVYY483 pKa = 10.43VSKK486 pKa = 9.05PTLDD490 pKa = 3.43VTPAADD496 pKa = 3.28MVVPYY501 pKa = 10.42LYY503 pKa = 10.56LIQPAAA509 pKa = 3.62
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84MLAFAAAAAVLLTAACGGGDD24 pKa = 3.43EE25 pKa = 4.61AATGGEE31 pKa = 4.57TTGSTASLVLGQTDD45 pKa = 3.45IPASFDD51 pKa = 3.34VLGYY55 pKa = 11.16ASGTKK60 pKa = 7.66TQYY63 pKa = 8.72FTAVYY68 pKa = 7.78DD69 pKa = 3.62TLLRR73 pKa = 11.84QDD75 pKa = 3.17GSGAIVPGLATDD87 pKa = 3.82WAYY90 pKa = 11.21DD91 pKa = 3.53DD92 pKa = 3.87TRR94 pKa = 11.84TVLTLTLRR102 pKa = 11.84EE103 pKa = 4.29GVTFTDD109 pKa = 5.54GVALDD114 pKa = 4.14ADD116 pKa = 3.52AVVANIEE123 pKa = 4.18HH124 pKa = 6.36FRR126 pKa = 11.84GSSTPDD132 pKa = 3.32LSNAAYY138 pKa = 9.39IDD140 pKa = 3.92TVSAVDD146 pKa = 3.36ATTVEE151 pKa = 4.21ITLTEE156 pKa = 4.83PDD158 pKa = 3.86PMMLTWLTQPLGYY171 pKa = 9.47MSSPEE176 pKa = 3.7SWEE179 pKa = 4.17NEE181 pKa = 3.84DD182 pKa = 4.01VATNPVGSGPYY193 pKa = 10.27VLDD196 pKa = 3.51TEE198 pKa = 4.46ATVVGSTYY206 pKa = 10.93VFTKK210 pKa = 10.82NPDD213 pKa = 3.09YY214 pKa = 10.8WDD216 pKa = 5.22DD217 pKa = 3.71SLQLFDD223 pKa = 4.51TLTMNYY229 pKa = 9.97YY230 pKa = 10.83ADD232 pKa = 3.99QSALLNALQGGQVDD246 pKa = 4.11AAPFNQFSALEE257 pKa = 3.99QVEE260 pKa = 4.22AAGYY264 pKa = 7.96EE265 pKa = 4.66SNVSRR270 pKa = 11.84LDD272 pKa = 3.32WEE274 pKa = 4.63GLMLLDD280 pKa = 5.23RR281 pKa = 11.84DD282 pKa = 4.19GALDD286 pKa = 3.69PALADD291 pKa = 3.24VRR293 pKa = 11.84VRR295 pKa = 11.84QAINHH300 pKa = 7.16AIAKK304 pKa = 10.06DD305 pKa = 3.99SILSAILLDD314 pKa = 4.17HH315 pKa = 6.48GTATSQIFGPSTDD328 pKa = 4.53GYY330 pKa = 11.33DD331 pKa = 3.43EE332 pKa = 5.13SLDD335 pKa = 3.51GYY337 pKa = 8.47YY338 pKa = 10.2TYY340 pKa = 11.35DD341 pKa = 3.61PDD343 pKa = 4.12KK344 pKa = 11.4AKK346 pKa = 10.99ALLAEE351 pKa = 4.66AGYY354 pKa = 11.36ADD356 pKa = 4.14GLEE359 pKa = 4.14LTMPSTSVIDD369 pKa = 3.76QALLTSIQQQLADD382 pKa = 3.48VGITVSYY389 pKa = 10.34TDD391 pKa = 3.15VGTNFISDD399 pKa = 3.86LMGAKK404 pKa = 9.97YY405 pKa = 10.3GSSWMRR411 pKa = 11.84LASSSDD417 pKa = 2.77WQTATFALTPDD428 pKa = 3.25ALFNSFRR435 pKa = 11.84TQRR438 pKa = 11.84PEE440 pKa = 3.25VDD442 pKa = 3.46ALLEE446 pKa = 4.2TVQKK450 pKa = 9.77GTEE453 pKa = 3.98EE454 pKa = 3.91EE455 pKa = 3.88AAAAAQEE462 pKa = 4.06LNRR465 pKa = 11.84YY466 pKa = 7.53VVEE469 pKa = 4.35EE470 pKa = 3.78AWFAPFYY477 pKa = 10.73FVDD480 pKa = 3.37NVYY483 pKa = 10.43VSKK486 pKa = 9.05PTLDD490 pKa = 3.43VTPAADD496 pKa = 3.28MVVPYY501 pKa = 10.42LYY503 pKa = 10.56LIQPAAA509 pKa = 3.62
Molecular weight: 54.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A542UW40|A0A542UW40_9CELL NADH-quinone oxidoreductase subunit C OS=Cellulomonas sp. SLBN-39 OX=2768446 GN=nuoC PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1256622 |
30 |
2247 |
338.8 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.197 ± 0.064 | 0.542 ± 0.01 |
6.676 ± 0.035 | 4.884 ± 0.043 |
2.262 ± 0.024 | 9.517 ± 0.041 |
2.187 ± 0.02 | 2.224 ± 0.028 |
1.102 ± 0.025 | 10.288 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.451 ± 0.016 | 1.282 ± 0.02 |
6.289 ± 0.036 | 2.679 ± 0.021 |
8.007 ± 0.05 | 4.475 ± 0.033 |
6.727 ± 0.052 | 10.936 ± 0.056 |
1.577 ± 0.016 | 1.697 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
