
Ulvibacter sp. KK4
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Patiriisocius
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
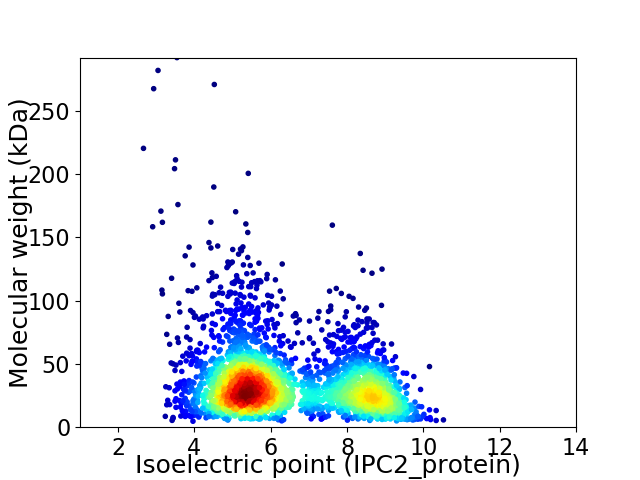
Virtual 2D-PAGE plot for 2972 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J4G032|A0A5J4G032_9FLAO 6-carboxy-5 6 7 8-tetrahydropterin synthase OS=Ulvibacter sp. KK4 OX=2494560 GN=ygcM PE=3 SV=1
MM1 pKa = 7.66KK2 pKa = 9.59NTFILLLIAISLWSCEE18 pKa = 3.74EE19 pKa = 4.15VIEE22 pKa = 4.86VEE24 pKa = 4.81LDD26 pKa = 3.19TTAPRR31 pKa = 11.84LVIDD35 pKa = 3.92ASINILLDD43 pKa = 4.34DD44 pKa = 4.74GSVTPKK50 pKa = 10.38IKK52 pKa = 10.63LSEE55 pKa = 3.96TAPFFGNEE63 pKa = 4.03VPPVTNAIVSITSDD77 pKa = 3.26NNEE80 pKa = 3.45VFMFEE85 pKa = 4.06HH86 pKa = 6.26QGDD89 pKa = 4.21GEE91 pKa = 4.52YY92 pKa = 7.84TTAIVPQLGVNYY104 pKa = 8.29TLKK107 pKa = 10.39IIHH110 pKa = 6.66NDD112 pKa = 2.94EE113 pKa = 4.52TYY115 pKa = 10.19TATTSLQTVVPLEE128 pKa = 4.07EE129 pKa = 3.93VTQNNEE135 pKa = 3.61GGFTGDD141 pKa = 3.46MIEE144 pKa = 4.25VEE146 pKa = 4.58ISFTDD151 pKa = 3.81PSNEE155 pKa = 3.41NNYY158 pKa = 10.11YY159 pKa = 10.34FFEE162 pKa = 4.55GLSDD166 pKa = 3.86RR167 pKa = 11.84GDD169 pKa = 3.68LRR171 pKa = 11.84DD172 pKa = 3.65SFSDD176 pKa = 3.33EE177 pKa = 4.1FFSGNTFQAIYY188 pKa = 9.62FADD191 pKa = 5.21DD192 pKa = 3.58IEE194 pKa = 6.14PDD196 pKa = 3.68DD197 pKa = 4.87VISFSLYY204 pKa = 10.33GVDD207 pKa = 3.15EE208 pKa = 3.97QFYY211 pKa = 10.44NFMFALLQQSSDD223 pKa = 3.01GGGPFEE229 pKa = 4.55TQPATIRR236 pKa = 11.84GNIINEE242 pKa = 4.43TNTDD246 pKa = 3.91NYY248 pKa = 9.38PLGYY252 pKa = 10.09FRR254 pKa = 11.84ISEE257 pKa = 4.16LSKK260 pKa = 10.38IVYY263 pKa = 8.58TVEE266 pKa = 3.62
MM1 pKa = 7.66KK2 pKa = 9.59NTFILLLIAISLWSCEE18 pKa = 3.74EE19 pKa = 4.15VIEE22 pKa = 4.86VEE24 pKa = 4.81LDD26 pKa = 3.19TTAPRR31 pKa = 11.84LVIDD35 pKa = 3.92ASINILLDD43 pKa = 4.34DD44 pKa = 4.74GSVTPKK50 pKa = 10.38IKK52 pKa = 10.63LSEE55 pKa = 3.96TAPFFGNEE63 pKa = 4.03VPPVTNAIVSITSDD77 pKa = 3.26NNEE80 pKa = 3.45VFMFEE85 pKa = 4.06HH86 pKa = 6.26QGDD89 pKa = 4.21GEE91 pKa = 4.52YY92 pKa = 7.84TTAIVPQLGVNYY104 pKa = 8.29TLKK107 pKa = 10.39IIHH110 pKa = 6.66NDD112 pKa = 2.94EE113 pKa = 4.52TYY115 pKa = 10.19TATTSLQTVVPLEE128 pKa = 4.07EE129 pKa = 3.93VTQNNEE135 pKa = 3.61GGFTGDD141 pKa = 3.46MIEE144 pKa = 4.25VEE146 pKa = 4.58ISFTDD151 pKa = 3.81PSNEE155 pKa = 3.41NNYY158 pKa = 10.11YY159 pKa = 10.34FFEE162 pKa = 4.55GLSDD166 pKa = 3.86RR167 pKa = 11.84GDD169 pKa = 3.68LRR171 pKa = 11.84DD172 pKa = 3.65SFSDD176 pKa = 3.33EE177 pKa = 4.1FFSGNTFQAIYY188 pKa = 9.62FADD191 pKa = 5.21DD192 pKa = 3.58IEE194 pKa = 6.14PDD196 pKa = 3.68DD197 pKa = 4.87VISFSLYY204 pKa = 10.33GVDD207 pKa = 3.15EE208 pKa = 3.97QFYY211 pKa = 10.44NFMFALLQQSSDD223 pKa = 3.01GGGPFEE229 pKa = 4.55TQPATIRR236 pKa = 11.84GNIINEE242 pKa = 4.43TNTDD246 pKa = 3.91NYY248 pKa = 9.38PLGYY252 pKa = 10.09FRR254 pKa = 11.84ISEE257 pKa = 4.16LSKK260 pKa = 10.38IVYY263 pKa = 8.58TVEE266 pKa = 3.62
Molecular weight: 29.72 kDa
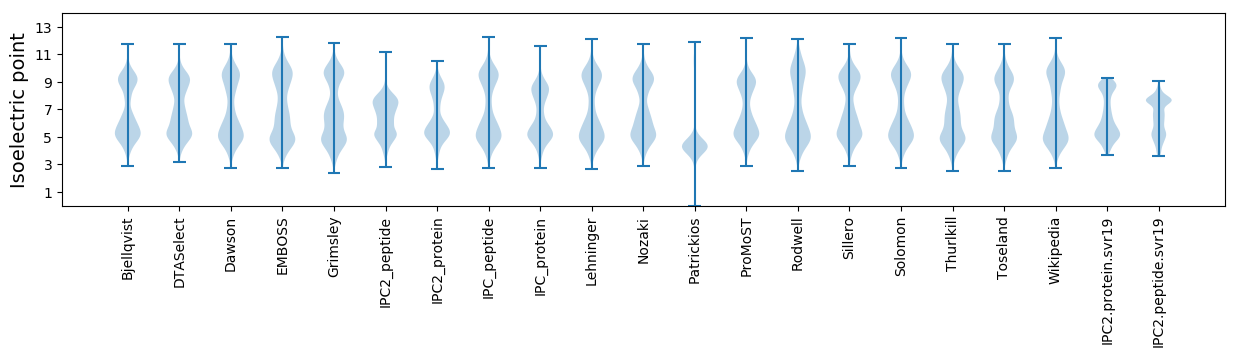
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J4FUW5|A0A5J4FUW5_9FLAO Uncharacterized protein OS=Ulvibacter sp. KK4 OX=2494560 GN=ULMS_03420 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.02RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGKK48 pKa = 6.48MTMRR52 pKa = 11.84YY53 pKa = 9.3IGGGHH58 pKa = 5.85KK59 pKa = 9.86KK60 pKa = 9.7KK61 pKa = 10.84YY62 pKa = 10.14RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.78RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 11.03AGVPATVATIEE82 pKa = 4.01YY83 pKa = 9.78DD84 pKa = 3.51PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 9.66QDD98 pKa = 3.47GEE100 pKa = 4.05KK101 pKa = 10.22RR102 pKa = 11.84YY103 pKa = 10.72VIAQNGLQVGQNIVSGEE120 pKa = 3.93QTTPEE125 pKa = 3.97IGNAMVLANVPLGTIISCIEE145 pKa = 3.75LRR147 pKa = 11.84PGQGAVLARR156 pKa = 11.84SAGAFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.05TRR182 pKa = 11.84LILVTCMATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.44GFRR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 9.1TKK262 pKa = 10.18QSNKK266 pKa = 10.2YY267 pKa = 9.54IIEE270 pKa = 4.05RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.02RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGKK48 pKa = 6.48MTMRR52 pKa = 11.84YY53 pKa = 9.3IGGGHH58 pKa = 5.85KK59 pKa = 9.86KK60 pKa = 9.7KK61 pKa = 10.84YY62 pKa = 10.14RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.78RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 11.03AGVPATVATIEE82 pKa = 4.01YY83 pKa = 9.78DD84 pKa = 3.51PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 9.66QDD98 pKa = 3.47GEE100 pKa = 4.05KK101 pKa = 10.22RR102 pKa = 11.84YY103 pKa = 10.72VIAQNGLQVGQNIVSGEE120 pKa = 3.93QTTPEE125 pKa = 3.97IGNAMVLANVPLGTIISCIEE145 pKa = 3.75LRR147 pKa = 11.84PGQGAVLARR156 pKa = 11.84SAGAFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.05TRR182 pKa = 11.84LILVTCMATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.44GFRR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 9.1TKK262 pKa = 10.18QSNKK266 pKa = 10.2YY267 pKa = 9.54IIEE270 pKa = 4.05RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989612 |
42 |
3042 |
333.0 |
37.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.585 ± 0.042 | 0.725 ± 0.02 |
5.714 ± 0.037 | 6.645 ± 0.047 |
5.185 ± 0.036 | 6.561 ± 0.068 |
1.644 ± 0.022 | 8.103 ± 0.043 |
7.572 ± 0.081 | 8.946 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.024 | 6.43 ± 0.057 |
3.336 ± 0.028 | 3.307 ± 0.026 |
3.349 ± 0.034 | 6.406 ± 0.038 |
6.198 ± 0.062 | 6.248 ± 0.036 |
0.954 ± 0.015 | 3.837 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
