
Trichodysplasia spinulosa-associated polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Human polyomavirus 8
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
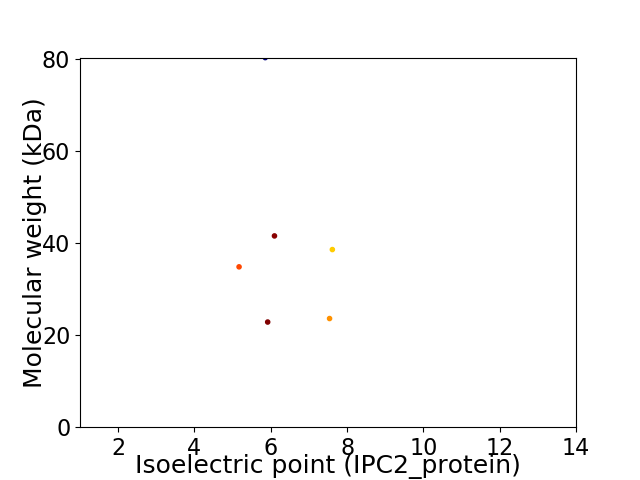
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2ESL6|E2ESL6_9POLY Minor capsid protein VP2 OS=Trichodysplasia spinulosa-associated polyomavirus OX=862909 GN=VP3 PE=3 SV=1
MM1 pKa = 7.47GGVISLFFDD10 pKa = 4.06IIEE13 pKa = 4.36ISTEE17 pKa = 3.59LSAATGFAVDD27 pKa = 3.91AVLAGEE33 pKa = 4.1AAAAVEE39 pKa = 4.28VEE41 pKa = 4.43VMGLMTVEE49 pKa = 4.33GLSASEE55 pKa = 4.01ALGTLGLTMEE65 pKa = 5.06NFSLMHH71 pKa = 7.14ALPGMLSEE79 pKa = 4.54AVGIGTLFQTISGASGLVAAGIRR102 pKa = 11.84YY103 pKa = 9.05GYY105 pKa = 10.05ARR107 pKa = 11.84EE108 pKa = 3.77VSIVNRR114 pKa = 11.84NISQMALQVWRR125 pKa = 11.84PWDD128 pKa = 3.54YY129 pKa = 11.83YY130 pKa = 11.28DD131 pKa = 3.81ILFPGVQTFAHH142 pKa = 5.91YY143 pKa = 11.18LNVLDD148 pKa = 4.88HH149 pKa = 6.52WASSLIHH156 pKa = 5.26TVSRR160 pKa = 11.84YY161 pKa = 7.19VWDD164 pKa = 5.56AILHH168 pKa = 5.7EE169 pKa = 4.76GRR171 pKa = 11.84HH172 pKa = 5.16QIGHH176 pKa = 7.33ASRR179 pKa = 11.84EE180 pKa = 3.95LMIRR184 pKa = 11.84GTNHH188 pKa = 5.86FQDD191 pKa = 3.52LMARR195 pKa = 11.84LIEE198 pKa = 3.94NSRR201 pKa = 11.84WVLTTGPSNIYY212 pKa = 10.14SHH214 pKa = 7.0LEE216 pKa = 3.56SYY218 pKa = 10.95YY219 pKa = 10.43RR220 pKa = 11.84DD221 pKa = 3.62LPGISPPQARR231 pKa = 11.84DD232 pKa = 3.15LYY234 pKa = 11.02RR235 pKa = 11.84RR236 pKa = 11.84LQEE239 pKa = 4.75KK240 pKa = 10.39IPDD243 pKa = 3.97RR244 pKa = 11.84YY245 pKa = 10.12QLEE248 pKa = 4.17AATDD252 pKa = 3.61EE253 pKa = 4.27SAEE256 pKa = 4.17VIEE259 pKa = 5.11TYY261 pKa = 10.25SAPGGAHH268 pKa = 6.07QRR270 pKa = 11.84VCPDD274 pKa = 2.28WMLPLVLGLYY284 pKa = 10.89GDD286 pKa = 3.82ITPTFGYY293 pKa = 9.37YY294 pKa = 10.07LRR296 pKa = 11.84EE297 pKa = 4.24EE298 pKa = 4.19EE299 pKa = 5.83RR300 pKa = 11.84EE301 pKa = 4.08DD302 pKa = 4.01GPQKK306 pKa = 9.98KK307 pKa = 9.09RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84MRR312 pKa = 3.59
MM1 pKa = 7.47GGVISLFFDD10 pKa = 4.06IIEE13 pKa = 4.36ISTEE17 pKa = 3.59LSAATGFAVDD27 pKa = 3.91AVLAGEE33 pKa = 4.1AAAAVEE39 pKa = 4.28VEE41 pKa = 4.43VMGLMTVEE49 pKa = 4.33GLSASEE55 pKa = 4.01ALGTLGLTMEE65 pKa = 5.06NFSLMHH71 pKa = 7.14ALPGMLSEE79 pKa = 4.54AVGIGTLFQTISGASGLVAAGIRR102 pKa = 11.84YY103 pKa = 9.05GYY105 pKa = 10.05ARR107 pKa = 11.84EE108 pKa = 3.77VSIVNRR114 pKa = 11.84NISQMALQVWRR125 pKa = 11.84PWDD128 pKa = 3.54YY129 pKa = 11.83YY130 pKa = 11.28DD131 pKa = 3.81ILFPGVQTFAHH142 pKa = 5.91YY143 pKa = 11.18LNVLDD148 pKa = 4.88HH149 pKa = 6.52WASSLIHH156 pKa = 5.26TVSRR160 pKa = 11.84YY161 pKa = 7.19VWDD164 pKa = 5.56AILHH168 pKa = 5.7EE169 pKa = 4.76GRR171 pKa = 11.84HH172 pKa = 5.16QIGHH176 pKa = 7.33ASRR179 pKa = 11.84EE180 pKa = 3.95LMIRR184 pKa = 11.84GTNHH188 pKa = 5.86FQDD191 pKa = 3.52LMARR195 pKa = 11.84LIEE198 pKa = 3.94NSRR201 pKa = 11.84WVLTTGPSNIYY212 pKa = 10.14SHH214 pKa = 7.0LEE216 pKa = 3.56SYY218 pKa = 10.95YY219 pKa = 10.43RR220 pKa = 11.84DD221 pKa = 3.62LPGISPPQARR231 pKa = 11.84DD232 pKa = 3.15LYY234 pKa = 11.02RR235 pKa = 11.84RR236 pKa = 11.84LQEE239 pKa = 4.75KK240 pKa = 10.39IPDD243 pKa = 3.97RR244 pKa = 11.84YY245 pKa = 10.12QLEE248 pKa = 4.17AATDD252 pKa = 3.61EE253 pKa = 4.27SAEE256 pKa = 4.17VIEE259 pKa = 5.11TYY261 pKa = 10.25SAPGGAHH268 pKa = 6.07QRR270 pKa = 11.84VCPDD274 pKa = 2.28WMLPLVLGLYY284 pKa = 10.89GDD286 pKa = 3.82ITPTFGYY293 pKa = 9.37YY294 pKa = 10.07LRR296 pKa = 11.84EE297 pKa = 4.24EE298 pKa = 4.19EE299 pKa = 5.83RR300 pKa = 11.84EE301 pKa = 4.08DD302 pKa = 4.01GPQKK306 pKa = 9.98KK307 pKa = 9.09RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84MRR312 pKa = 3.59
Molecular weight: 34.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2ESL9|E2ESL9_9POLY Isoform of E2ESL8 Small T antigen OS=Trichodysplasia spinulosa-associated polyomavirus OX=862909 GN=T PE=4 SV=1
MM1 pKa = 8.02DD2 pKa = 4.71KK3 pKa = 10.5FLSRR7 pKa = 11.84EE8 pKa = 3.83EE9 pKa = 4.2SLEE12 pKa = 4.7LMDD15 pKa = 5.59LLQIPRR21 pKa = 11.84HH22 pKa = 5.91CYY24 pKa = 10.76GNFALMKK31 pKa = 9.53INHH34 pKa = 6.61KK35 pKa = 10.58KK36 pKa = 9.86MSLKK40 pKa = 9.98YY41 pKa = 10.25HH42 pKa = 6.86PDD44 pKa = 3.2KK45 pKa = 11.6GGDD48 pKa = 3.62PEE50 pKa = 4.85KK51 pKa = 10.3MSRR54 pKa = 11.84LNQLWQKK61 pKa = 9.89LQEE64 pKa = 4.63GIYY67 pKa = 10.02NARR70 pKa = 11.84QEE72 pKa = 4.44FPTSFSSQVGSWYY85 pKa = 9.79WEE87 pKa = 3.89ANLISLKK94 pKa = 10.63EE95 pKa = 3.88YY96 pKa = 10.45FGKK99 pKa = 10.4KK100 pKa = 9.98KK101 pKa = 10.51YY102 pKa = 10.12DD103 pKa = 3.56EE104 pKa = 4.47NVIKK108 pKa = 10.21HH109 pKa = 5.71WPQCAEE115 pKa = 4.2KK116 pKa = 10.57ALKK119 pKa = 9.61EE120 pKa = 4.47CKK122 pKa = 10.23CLTCKK127 pKa = 10.23IGLQHH132 pKa = 6.61HH133 pKa = 6.95VYY135 pKa = 10.61KK136 pKa = 10.85QMHH139 pKa = 5.09QKK141 pKa = 10.22KK142 pKa = 8.66CVVWGEE148 pKa = 4.11CFCYY152 pKa = 10.02KK153 pKa = 10.3CYY155 pKa = 10.41CAWFGEE161 pKa = 4.26DD162 pKa = 5.38LYY164 pKa = 11.69CLDD167 pKa = 5.53SLWAWSCIVGEE178 pKa = 4.27VDD180 pKa = 3.83FHH182 pKa = 7.3LVNLYY187 pKa = 10.79LRR189 pKa = 11.84VNQGFNWVFPFSMMFQPRR207 pKa = 11.84MEE209 pKa = 4.89EE210 pKa = 3.67IYY212 pKa = 10.8LPMGTPPGPAGGKK225 pKa = 10.14ASIKK229 pKa = 10.89NGTTCLTPCRR239 pKa = 11.84TQISSAMNPPFPLMNLDD256 pKa = 3.51LQAPLRR262 pKa = 11.84DD263 pKa = 3.61PLLNLARR270 pKa = 11.84RR271 pKa = 11.84IQEE274 pKa = 4.11EE275 pKa = 4.47EE276 pKa = 4.13EE277 pKa = 4.55LPHH280 pKa = 6.72QRR282 pKa = 11.84TPPAAPRR289 pKa = 11.84APSLPPPQSQKK300 pKa = 10.63NLSMTLSLMIFLICCGLFFLMLSIVIKK327 pKa = 10.42LYY329 pKa = 10.94HH330 pKa = 6.53LFF332 pKa = 4.02
MM1 pKa = 8.02DD2 pKa = 4.71KK3 pKa = 10.5FLSRR7 pKa = 11.84EE8 pKa = 3.83EE9 pKa = 4.2SLEE12 pKa = 4.7LMDD15 pKa = 5.59LLQIPRR21 pKa = 11.84HH22 pKa = 5.91CYY24 pKa = 10.76GNFALMKK31 pKa = 9.53INHH34 pKa = 6.61KK35 pKa = 10.58KK36 pKa = 9.86MSLKK40 pKa = 9.98YY41 pKa = 10.25HH42 pKa = 6.86PDD44 pKa = 3.2KK45 pKa = 11.6GGDD48 pKa = 3.62PEE50 pKa = 4.85KK51 pKa = 10.3MSRR54 pKa = 11.84LNQLWQKK61 pKa = 9.89LQEE64 pKa = 4.63GIYY67 pKa = 10.02NARR70 pKa = 11.84QEE72 pKa = 4.44FPTSFSSQVGSWYY85 pKa = 9.79WEE87 pKa = 3.89ANLISLKK94 pKa = 10.63EE95 pKa = 3.88YY96 pKa = 10.45FGKK99 pKa = 10.4KK100 pKa = 9.98KK101 pKa = 10.51YY102 pKa = 10.12DD103 pKa = 3.56EE104 pKa = 4.47NVIKK108 pKa = 10.21HH109 pKa = 5.71WPQCAEE115 pKa = 4.2KK116 pKa = 10.57ALKK119 pKa = 9.61EE120 pKa = 4.47CKK122 pKa = 10.23CLTCKK127 pKa = 10.23IGLQHH132 pKa = 6.61HH133 pKa = 6.95VYY135 pKa = 10.61KK136 pKa = 10.85QMHH139 pKa = 5.09QKK141 pKa = 10.22KK142 pKa = 8.66CVVWGEE148 pKa = 4.11CFCYY152 pKa = 10.02KK153 pKa = 10.3CYY155 pKa = 10.41CAWFGEE161 pKa = 4.26DD162 pKa = 5.38LYY164 pKa = 11.69CLDD167 pKa = 5.53SLWAWSCIVGEE178 pKa = 4.27VDD180 pKa = 3.83FHH182 pKa = 7.3LVNLYY187 pKa = 10.79LRR189 pKa = 11.84VNQGFNWVFPFSMMFQPRR207 pKa = 11.84MEE209 pKa = 4.89EE210 pKa = 3.67IYY212 pKa = 10.8LPMGTPPGPAGGKK225 pKa = 10.14ASIKK229 pKa = 10.89NGTTCLTPCRR239 pKa = 11.84TQISSAMNPPFPLMNLDD256 pKa = 3.51LQAPLRR262 pKa = 11.84DD263 pKa = 3.61PLLNLARR270 pKa = 11.84RR271 pKa = 11.84IQEE274 pKa = 4.11EE275 pKa = 4.47EE276 pKa = 4.13EE277 pKa = 4.55LPHH280 pKa = 6.72QRR282 pKa = 11.84TPPAAPRR289 pKa = 11.84APSLPPPQSQKK300 pKa = 10.63NLSMTLSLMIFLICCGLFFLMLSIVIKK327 pKa = 10.42LYY329 pKa = 10.94HH330 pKa = 6.53LFF332 pKa = 4.02
Molecular weight: 38.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2108 |
194 |
697 |
351.3 |
40.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.981 ± 0.833 | 2.846 ± 0.656 |
4.981 ± 0.408 | 6.784 ± 0.514 |
4.317 ± 0.73 | 6.12 ± 1.048 |
2.799 ± 0.355 | 4.412 ± 0.441 |
6.452 ± 1.418 | 10.389 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.304 | 4.507 ± 0.623 |
6.025 ± 0.669 | 4.269 ± 0.273 |
4.744 ± 0.801 | 6.357 ± 0.412 |
4.934 ± 0.764 | 5.55 ± 0.576 |
1.945 ± 0.426 | 4.364 ± 0.531 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
