
Sporobacter termitidis DSM 10068
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Sporobacter; Sporobacter termitidis
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
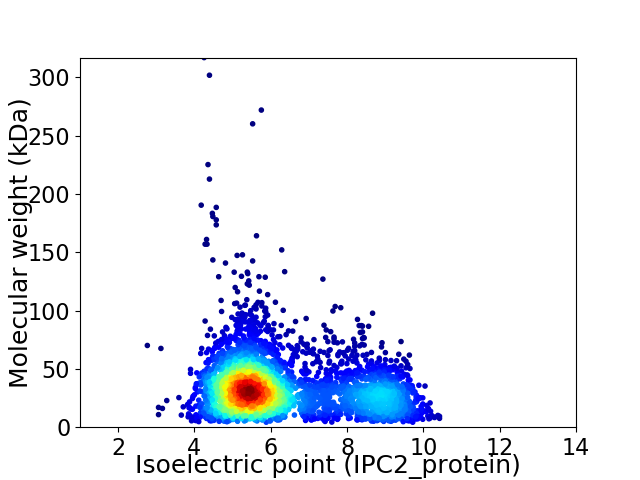
Virtual 2D-PAGE plot for 3771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5WIX7|A0A1M5WIX7_9FIRM Cysteine desulfurase OS=Sporobacter termitidis DSM 10068 OX=1123282 GN=SAMN02745823_01303 PE=3 SV=1
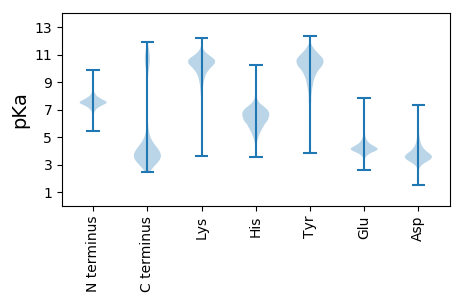
MM1 pKa = 7.2TPEE4 pKa = 4.19ASAPSPTPDD13 pKa = 4.19AAPLPPGAFAVSSGNIHH30 pKa = 7.75DD31 pKa = 4.76YY32 pKa = 11.26LPDD35 pKa = 5.08LIVGKK40 pKa = 9.18PVSAYY45 pKa = 10.71ALLPCLDD52 pKa = 3.79SFSRR56 pKa = 11.84GTWSEE61 pKa = 4.23LEE63 pKa = 4.12TAYY66 pKa = 10.47GGPDD70 pKa = 3.27GTDD73 pKa = 2.14WWSPLLTALNDD84 pKa = 3.53SAAGDD89 pKa = 3.94DD90 pKa = 3.76QEE92 pKa = 4.39MRR94 pKa = 11.84DD95 pKa = 4.04YY96 pKa = 11.31YY97 pKa = 10.96LGRR100 pKa = 11.84AYY102 pKa = 9.5LTSDD106 pKa = 3.38GAYY109 pKa = 10.24SEE111 pKa = 5.46GLDD114 pKa = 4.7GPLLAQWEE122 pKa = 4.52ADD124 pKa = 3.19STAYY128 pKa = 10.01NACLSGRR135 pKa = 11.84FSADD139 pKa = 2.49EE140 pKa = 3.9ATALRR145 pKa = 11.84QALTFAVGSDD155 pKa = 3.12GNAYY159 pKa = 10.3GLYY162 pKa = 10.54LPGAARR168 pKa = 11.84AIYY171 pKa = 9.83LDD173 pKa = 4.39KK174 pKa = 11.45YY175 pKa = 10.63PIDD178 pKa = 3.74FPFGFEE184 pKa = 3.69LRR186 pKa = 11.84EE187 pKa = 3.84NSRR190 pKa = 11.84SSFRR194 pKa = 11.84AEE196 pKa = 3.84SFGPGGVVEE205 pKa = 4.47SDD207 pKa = 3.41GLQVTYY213 pKa = 8.41LTPEE217 pKa = 4.5DD218 pKa = 4.07GVFSVTTIRR227 pKa = 11.84TVKK230 pKa = 9.55EE231 pKa = 4.01GCGIWGVAVGQPEE244 pKa = 4.07RR245 pKa = 11.84VLLNSSIPLKK255 pKa = 10.81KK256 pKa = 9.28IDD258 pKa = 3.71SLSYY262 pKa = 11.31DD263 pKa = 3.29DD264 pKa = 5.44AAWFGKK270 pKa = 9.7CDD272 pKa = 3.06RR273 pKa = 11.84VYY275 pKa = 11.41AFLPEE280 pKa = 3.98EE281 pKa = 4.04STIAVVFAVKK291 pKa = 10.53DD292 pKa = 3.61DD293 pKa = 3.67VVYY296 pKa = 10.36GIEE299 pKa = 5.32LINGLDD305 pKa = 3.48GQMYY309 pKa = 10.42
MM1 pKa = 7.2TPEE4 pKa = 4.19ASAPSPTPDD13 pKa = 4.19AAPLPPGAFAVSSGNIHH30 pKa = 7.75DD31 pKa = 4.76YY32 pKa = 11.26LPDD35 pKa = 5.08LIVGKK40 pKa = 9.18PVSAYY45 pKa = 10.71ALLPCLDD52 pKa = 3.79SFSRR56 pKa = 11.84GTWSEE61 pKa = 4.23LEE63 pKa = 4.12TAYY66 pKa = 10.47GGPDD70 pKa = 3.27GTDD73 pKa = 2.14WWSPLLTALNDD84 pKa = 3.53SAAGDD89 pKa = 3.94DD90 pKa = 3.76QEE92 pKa = 4.39MRR94 pKa = 11.84DD95 pKa = 4.04YY96 pKa = 11.31YY97 pKa = 10.96LGRR100 pKa = 11.84AYY102 pKa = 9.5LTSDD106 pKa = 3.38GAYY109 pKa = 10.24SEE111 pKa = 5.46GLDD114 pKa = 4.7GPLLAQWEE122 pKa = 4.52ADD124 pKa = 3.19STAYY128 pKa = 10.01NACLSGRR135 pKa = 11.84FSADD139 pKa = 2.49EE140 pKa = 3.9ATALRR145 pKa = 11.84QALTFAVGSDD155 pKa = 3.12GNAYY159 pKa = 10.3GLYY162 pKa = 10.54LPGAARR168 pKa = 11.84AIYY171 pKa = 9.83LDD173 pKa = 4.39KK174 pKa = 11.45YY175 pKa = 10.63PIDD178 pKa = 3.74FPFGFEE184 pKa = 3.69LRR186 pKa = 11.84EE187 pKa = 3.84NSRR190 pKa = 11.84SSFRR194 pKa = 11.84AEE196 pKa = 3.84SFGPGGVVEE205 pKa = 4.47SDD207 pKa = 3.41GLQVTYY213 pKa = 8.41LTPEE217 pKa = 4.5DD218 pKa = 4.07GVFSVTTIRR227 pKa = 11.84TVKK230 pKa = 9.55EE231 pKa = 4.01GCGIWGVAVGQPEE244 pKa = 4.07RR245 pKa = 11.84VLLNSSIPLKK255 pKa = 10.81KK256 pKa = 9.28IDD258 pKa = 3.71SLSYY262 pKa = 11.31DD263 pKa = 3.29DD264 pKa = 5.44AAWFGKK270 pKa = 9.7CDD272 pKa = 3.06RR273 pKa = 11.84VYY275 pKa = 11.41AFLPEE280 pKa = 3.98EE281 pKa = 4.04STIAVVFAVKK291 pKa = 10.53DD292 pKa = 3.61DD293 pKa = 3.67VVYY296 pKa = 10.36GIEE299 pKa = 5.32LINGLDD305 pKa = 3.48GQMYY309 pKa = 10.42
Molecular weight: 33.12 kDa
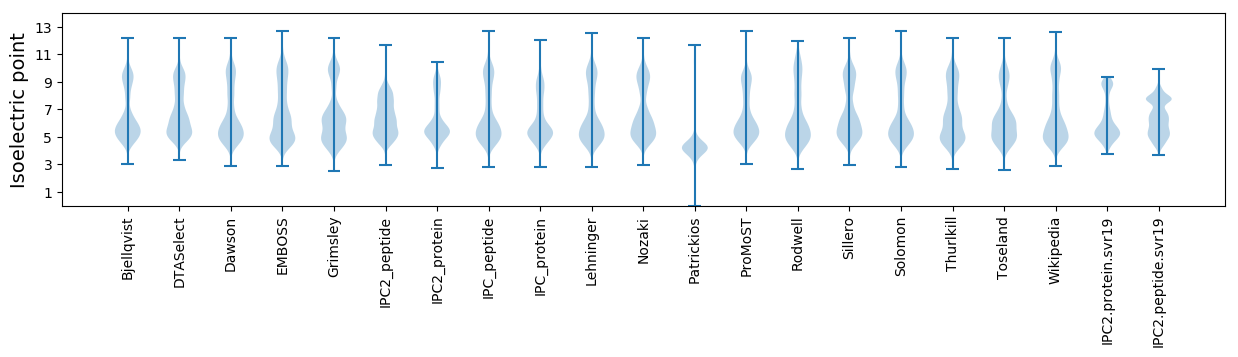
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5UMZ3|A0A1M5UMZ3_9FIRM Putative adhesin OS=Sporobacter termitidis DSM 10068 OX=1123282 GN=SAMN02745823_00594 PE=4 SV=1
MM1 pKa = 7.46ANEE4 pKa = 3.75RR5 pKa = 11.84DD6 pKa = 3.82GRR8 pKa = 11.84DD9 pKa = 3.48GRR11 pKa = 11.84DD12 pKa = 3.39GRR14 pKa = 11.84GHH16 pKa = 4.96GRR18 pKa = 11.84KK19 pKa = 8.98RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.04VCQFCVDD29 pKa = 3.12KK30 pKa = 11.13AQNIDD35 pKa = 3.64YY36 pKa = 10.57KK37 pKa = 10.5DD38 pKa = 3.14TAKK41 pKa = 10.09IRR43 pKa = 11.84RR44 pKa = 11.84YY45 pKa = 9.1VSEE48 pKa = 3.99RR49 pKa = 11.84SKK51 pKa = 10.6ILPRR55 pKa = 11.84RR56 pKa = 11.84TTGICAQHH64 pKa = 5.68QRR66 pKa = 11.84QLMVAIKK73 pKa = 10.13RR74 pKa = 11.84ARR76 pKa = 11.84QIALLPYY83 pKa = 9.27VTDD86 pKa = 3.53
MM1 pKa = 7.46ANEE4 pKa = 3.75RR5 pKa = 11.84DD6 pKa = 3.82GRR8 pKa = 11.84DD9 pKa = 3.48GRR11 pKa = 11.84DD12 pKa = 3.39GRR14 pKa = 11.84GHH16 pKa = 4.96GRR18 pKa = 11.84KK19 pKa = 8.98RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.04VCQFCVDD29 pKa = 3.12KK30 pKa = 11.13AQNIDD35 pKa = 3.64YY36 pKa = 10.57KK37 pKa = 10.5DD38 pKa = 3.14TAKK41 pKa = 10.09IRR43 pKa = 11.84RR44 pKa = 11.84YY45 pKa = 9.1VSEE48 pKa = 3.99RR49 pKa = 11.84SKK51 pKa = 10.6ILPRR55 pKa = 11.84RR56 pKa = 11.84TTGICAQHH64 pKa = 5.68QRR66 pKa = 11.84QLMVAIKK73 pKa = 10.13RR74 pKa = 11.84ARR76 pKa = 11.84QIALLPYY83 pKa = 9.27VTDD86 pKa = 3.53
Molecular weight: 10.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1206611 |
39 |
3160 |
320.0 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.791 ± 0.046 | 1.416 ± 0.017 |
5.661 ± 0.033 | 5.942 ± 0.042 |
4.032 ± 0.028 | 8.148 ± 0.051 |
1.604 ± 0.016 | 6.571 ± 0.034 |
5.392 ± 0.032 | 9.643 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.687 ± 0.023 | 3.692 ± 0.029 |
4.059 ± 0.026 | 2.779 ± 0.02 |
5.086 ± 0.041 | 6.062 ± 0.038 |
5.731 ± 0.046 | 7.234 ± 0.034 |
0.895 ± 0.013 | 3.574 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
