
Actinidia seed borne latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Prunevirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
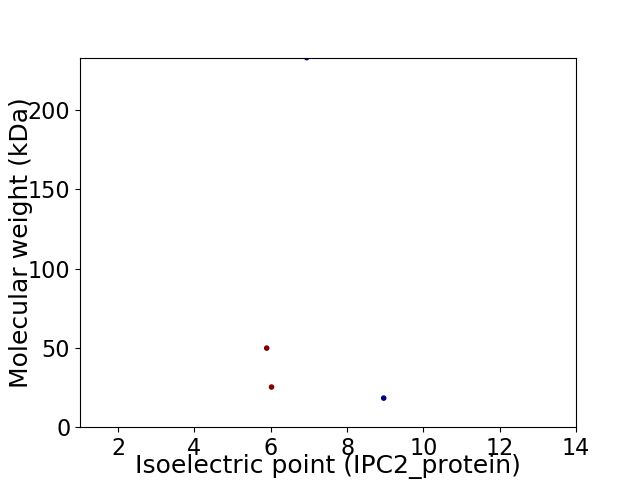
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
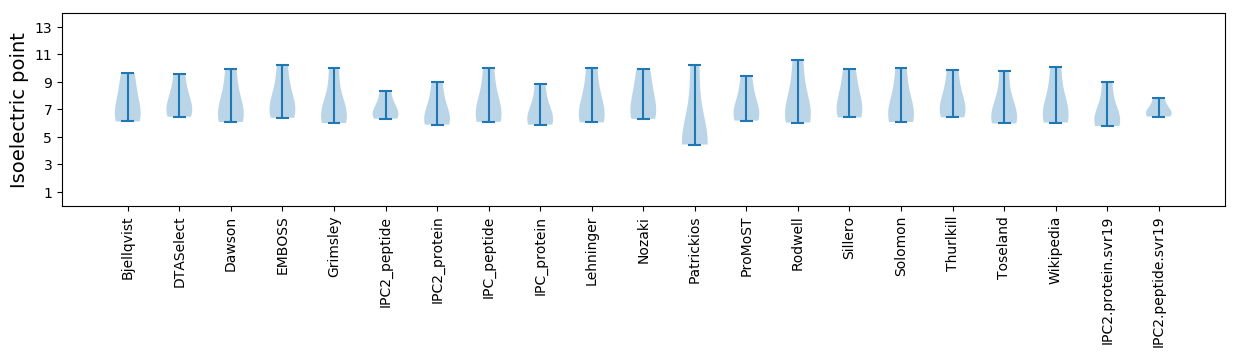
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L0V1P5|A0A2L0V1P5_9VIRU Nucleic acid binding protein OS=Actinidia seed borne latent virus OX=2560282 PE=4 SV=1
MM1 pKa = 7.37SKK3 pKa = 10.24SIKK6 pKa = 10.21VNSIVNRR13 pKa = 11.84VNVDD17 pKa = 3.12KK18 pKa = 11.18SILGSSEE25 pKa = 3.36ISSLYY30 pKa = 8.58GSKK33 pKa = 10.34YY34 pKa = 10.37APLVFKK40 pKa = 10.98DD41 pKa = 4.01EE42 pKa = 4.14VKK44 pKa = 10.45MIVPGNILGGPIKK57 pKa = 10.51LQANILTKK65 pKa = 10.42EE66 pKa = 3.67RR67 pKa = 11.84LGIIRR72 pKa = 11.84GQKK75 pKa = 10.24FGGSNCAYY83 pKa = 9.69LHH85 pKa = 6.49LGFVPIVIQSLLVSGNEE102 pKa = 4.01LVKK105 pKa = 10.74GRR107 pKa = 11.84CSLVDD112 pKa = 3.82LSRR115 pKa = 11.84GSEE118 pKa = 3.86KK119 pKa = 10.3TGLIDD124 pKa = 3.85RR125 pKa = 11.84FNFKK129 pKa = 8.67FTKK132 pKa = 10.43NEE134 pKa = 3.49PFAAKK139 pKa = 9.83ILTINAPIDD148 pKa = 3.77INCDD152 pKa = 3.02TSINSIQILLEE163 pKa = 3.92LEE165 pKa = 5.07GIDD168 pKa = 3.36IRR170 pKa = 11.84TEE172 pKa = 3.72RR173 pKa = 11.84SVIAVVTGLSCVPTNSTIMLPGLKK197 pKa = 9.67RR198 pKa = 11.84EE199 pKa = 4.1TPKK202 pKa = 10.5WSICNVFNVPEE213 pKa = 4.03EE214 pKa = 4.25SEE216 pKa = 4.07EE217 pKa = 4.04DD218 pKa = 3.41NEE220 pKa = 4.82RR221 pKa = 11.84FNSLFNGANPKK232 pKa = 10.56LIDD235 pKa = 3.99LGKK238 pKa = 9.55DD239 pKa = 3.1TVLDD243 pKa = 3.65NGKK246 pKa = 9.84KK247 pKa = 9.91FGFWGPSVKK256 pKa = 9.7PVHH259 pKa = 6.55RR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 4.15LTTKK266 pKa = 10.76NIIKK270 pKa = 9.64EE271 pKa = 3.98QMSQVMSEE279 pKa = 4.33TVSNLNICEE288 pKa = 4.0EE289 pKa = 4.13RR290 pKa = 11.84KK291 pKa = 10.0KK292 pKa = 10.9LRR294 pKa = 11.84NCLEE298 pKa = 3.71RR299 pKa = 11.84SKK301 pKa = 10.92SVRR304 pKa = 11.84GNVSEE309 pKa = 4.22VGKK312 pKa = 10.84EE313 pKa = 3.78EE314 pKa = 4.25VPVRR318 pKa = 11.84RR319 pKa = 11.84SLRR322 pKa = 11.84VDD324 pKa = 3.03MEE326 pKa = 4.04EE327 pKa = 3.85LHH329 pKa = 7.4RR330 pKa = 11.84SGRR333 pKa = 11.84QDD335 pKa = 2.64ARR337 pKa = 11.84MEE339 pKa = 4.59GGQQWEE345 pKa = 4.82CEE347 pKa = 3.88ASKK350 pKa = 10.82YY351 pKa = 10.13KK352 pKa = 10.51HH353 pKa = 6.96DD354 pKa = 3.66GTEE357 pKa = 3.78VKK359 pKa = 10.42DD360 pKa = 3.79YY361 pKa = 11.27DD362 pKa = 3.9GSAEE366 pKa = 3.94SDD368 pKa = 2.71IRR370 pKa = 11.84EE371 pKa = 4.08YY372 pKa = 11.14CRR374 pKa = 11.84LLGGSRR380 pKa = 11.84NKK382 pKa = 10.66LSIDD386 pKa = 3.63YY387 pKa = 10.98SGLQCDD393 pKa = 3.3EE394 pKa = 5.11CEE396 pKa = 4.04WDD398 pKa = 3.57RR399 pKa = 11.84KK400 pKa = 10.33SDD402 pKa = 3.23INRR405 pKa = 11.84WEE407 pKa = 4.06FQSEE411 pKa = 4.25DD412 pKa = 3.3NSGNNEE418 pKa = 4.22DD419 pKa = 4.13LHH421 pKa = 7.29SNPSGSEE428 pKa = 4.0DD429 pKa = 3.6IIINAKK435 pKa = 10.36GGLQEE440 pKa = 4.25FCEE443 pKa = 4.33LRR445 pKa = 11.84PP446 pKa = 3.63
MM1 pKa = 7.37SKK3 pKa = 10.24SIKK6 pKa = 10.21VNSIVNRR13 pKa = 11.84VNVDD17 pKa = 3.12KK18 pKa = 11.18SILGSSEE25 pKa = 3.36ISSLYY30 pKa = 8.58GSKK33 pKa = 10.34YY34 pKa = 10.37APLVFKK40 pKa = 10.98DD41 pKa = 4.01EE42 pKa = 4.14VKK44 pKa = 10.45MIVPGNILGGPIKK57 pKa = 10.51LQANILTKK65 pKa = 10.42EE66 pKa = 3.67RR67 pKa = 11.84LGIIRR72 pKa = 11.84GQKK75 pKa = 10.24FGGSNCAYY83 pKa = 9.69LHH85 pKa = 6.49LGFVPIVIQSLLVSGNEE102 pKa = 4.01LVKK105 pKa = 10.74GRR107 pKa = 11.84CSLVDD112 pKa = 3.82LSRR115 pKa = 11.84GSEE118 pKa = 3.86KK119 pKa = 10.3TGLIDD124 pKa = 3.85RR125 pKa = 11.84FNFKK129 pKa = 8.67FTKK132 pKa = 10.43NEE134 pKa = 3.49PFAAKK139 pKa = 9.83ILTINAPIDD148 pKa = 3.77INCDD152 pKa = 3.02TSINSIQILLEE163 pKa = 3.92LEE165 pKa = 5.07GIDD168 pKa = 3.36IRR170 pKa = 11.84TEE172 pKa = 3.72RR173 pKa = 11.84SVIAVVTGLSCVPTNSTIMLPGLKK197 pKa = 9.67RR198 pKa = 11.84EE199 pKa = 4.1TPKK202 pKa = 10.5WSICNVFNVPEE213 pKa = 4.03EE214 pKa = 4.25SEE216 pKa = 4.07EE217 pKa = 4.04DD218 pKa = 3.41NEE220 pKa = 4.82RR221 pKa = 11.84FNSLFNGANPKK232 pKa = 10.56LIDD235 pKa = 3.99LGKK238 pKa = 9.55DD239 pKa = 3.1TVLDD243 pKa = 3.65NGKK246 pKa = 9.84KK247 pKa = 9.91FGFWGPSVKK256 pKa = 9.7PVHH259 pKa = 6.55RR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 4.15LTTKK266 pKa = 10.76NIIKK270 pKa = 9.64EE271 pKa = 3.98QMSQVMSEE279 pKa = 4.33TVSNLNICEE288 pKa = 4.0EE289 pKa = 4.13RR290 pKa = 11.84KK291 pKa = 10.0KK292 pKa = 10.9LRR294 pKa = 11.84NCLEE298 pKa = 3.71RR299 pKa = 11.84SKK301 pKa = 10.92SVRR304 pKa = 11.84GNVSEE309 pKa = 4.22VGKK312 pKa = 10.84EE313 pKa = 3.78EE314 pKa = 4.25VPVRR318 pKa = 11.84RR319 pKa = 11.84SLRR322 pKa = 11.84VDD324 pKa = 3.03MEE326 pKa = 4.04EE327 pKa = 3.85LHH329 pKa = 7.4RR330 pKa = 11.84SGRR333 pKa = 11.84QDD335 pKa = 2.64ARR337 pKa = 11.84MEE339 pKa = 4.59GGQQWEE345 pKa = 4.82CEE347 pKa = 3.88ASKK350 pKa = 10.82YY351 pKa = 10.13KK352 pKa = 10.51HH353 pKa = 6.96DD354 pKa = 3.66GTEE357 pKa = 3.78VKK359 pKa = 10.42DD360 pKa = 3.79YY361 pKa = 11.27DD362 pKa = 3.9GSAEE366 pKa = 3.94SDD368 pKa = 2.71IRR370 pKa = 11.84EE371 pKa = 4.08YY372 pKa = 11.14CRR374 pKa = 11.84LLGGSRR380 pKa = 11.84NKK382 pKa = 10.66LSIDD386 pKa = 3.63YY387 pKa = 10.98SGLQCDD393 pKa = 3.3EE394 pKa = 5.11CEE396 pKa = 4.04WDD398 pKa = 3.57RR399 pKa = 11.84KK400 pKa = 10.33SDD402 pKa = 3.23INRR405 pKa = 11.84WEE407 pKa = 4.06FQSEE411 pKa = 4.25DD412 pKa = 3.3NSGNNEE418 pKa = 4.22DD419 pKa = 4.13LHH421 pKa = 7.29SNPSGSEE428 pKa = 4.0DD429 pKa = 3.6IIINAKK435 pKa = 10.36GGLQEE440 pKa = 4.25FCEE443 pKa = 4.33LRR445 pKa = 11.84PP446 pKa = 3.63
Molecular weight: 49.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L0V1P9|A0A2L0V1P9_9VIRU Coat protein OS=Actinidia seed borne latent virus OX=2560282 PE=4 SV=1
MM1 pKa = 7.46EE2 pKa = 5.16KK3 pKa = 10.75AEE5 pKa = 4.21SFAGKK10 pKa = 8.31TIKK13 pKa = 10.51NWVALNKK20 pKa = 10.55ANIFLSLINSFGRR33 pKa = 11.84DD34 pKa = 2.65IGFKK38 pKa = 9.99IFMMYY43 pKa = 9.79KK44 pKa = 9.5CKK46 pKa = 10.46CEE48 pKa = 4.4KK49 pKa = 10.17EE50 pKa = 4.16YY51 pKa = 11.06KK52 pKa = 10.41DD53 pKa = 3.43QIRR56 pKa = 11.84YY57 pKa = 9.22QNMVNYY63 pKa = 9.66GQGKK67 pKa = 9.3SKK69 pKa = 11.0SAMKK73 pKa = 10.2RR74 pKa = 11.84RR75 pKa = 11.84AIKK78 pKa = 10.45VEE80 pKa = 3.61HH81 pKa = 6.99CYY83 pKa = 10.69KK84 pKa = 10.46CGKK87 pKa = 9.69FEE89 pKa = 6.07HH90 pKa = 7.43DD91 pKa = 3.35GHH93 pKa = 6.46CNKK96 pKa = 10.0NQTNSNHH103 pKa = 6.58EE104 pKa = 4.09YY105 pKa = 11.23LEE107 pKa = 3.98MFRR110 pKa = 11.84CGPIKK115 pKa = 10.58LKK117 pKa = 10.9AEE119 pKa = 3.63RR120 pKa = 11.84ALRR123 pKa = 11.84KK124 pKa = 10.1NSMVQMSCEE133 pKa = 3.75KK134 pKa = 9.83FGWMIKK140 pKa = 8.78MSKK143 pKa = 8.48EE144 pKa = 3.97LKK146 pKa = 9.94EE147 pKa = 4.05KK148 pKa = 10.78ANSGTAEE155 pKa = 3.98RR156 pKa = 11.84VV157 pKa = 3.17
MM1 pKa = 7.46EE2 pKa = 5.16KK3 pKa = 10.75AEE5 pKa = 4.21SFAGKK10 pKa = 8.31TIKK13 pKa = 10.51NWVALNKK20 pKa = 10.55ANIFLSLINSFGRR33 pKa = 11.84DD34 pKa = 2.65IGFKK38 pKa = 9.99IFMMYY43 pKa = 9.79KK44 pKa = 9.5CKK46 pKa = 10.46CEE48 pKa = 4.4KK49 pKa = 10.17EE50 pKa = 4.16YY51 pKa = 11.06KK52 pKa = 10.41DD53 pKa = 3.43QIRR56 pKa = 11.84YY57 pKa = 9.22QNMVNYY63 pKa = 9.66GQGKK67 pKa = 9.3SKK69 pKa = 11.0SAMKK73 pKa = 10.2RR74 pKa = 11.84RR75 pKa = 11.84AIKK78 pKa = 10.45VEE80 pKa = 3.61HH81 pKa = 6.99CYY83 pKa = 10.69KK84 pKa = 10.46CGKK87 pKa = 9.69FEE89 pKa = 6.07HH90 pKa = 7.43DD91 pKa = 3.35GHH93 pKa = 6.46CNKK96 pKa = 10.0NQTNSNHH103 pKa = 6.58EE104 pKa = 4.09YY105 pKa = 11.23LEE107 pKa = 3.98MFRR110 pKa = 11.84CGPIKK115 pKa = 10.58LKK117 pKa = 10.9AEE119 pKa = 3.63RR120 pKa = 11.84ALRR123 pKa = 11.84KK124 pKa = 10.1NSMVQMSCEE133 pKa = 3.75KK134 pKa = 9.83FGWMIKK140 pKa = 8.78MSKK143 pKa = 8.48EE144 pKa = 3.97LKK146 pKa = 9.94EE147 pKa = 4.05KK148 pKa = 10.78ANSGTAEE155 pKa = 3.98RR156 pKa = 11.84VV157 pKa = 3.17
Molecular weight: 18.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2827 |
157 |
2002 |
706.8 |
81.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.174 ± 0.816 | 2.335 ± 0.535 |
5.766 ± 0.805 | 8.56 ± 0.648 |
5.624 ± 0.904 | 5.341 ± 0.929 |
2.299 ± 0.47 | 6.969 ± 0.344 |
9.268 ± 1.519 | 7.853 ± 0.672 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.042 ± 0.782 | 6.155 ± 0.748 |
2.688 ± 0.452 | 2.618 ± 0.267 |
5.624 ± 0.426 | 7.676 ± 0.804 |
3.714 ± 0.662 | 6.19 ± 0.587 |
1.097 ± 0.074 | 3.007 ± 0.514 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
