
Shinella sp. MEC087
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Shinella
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
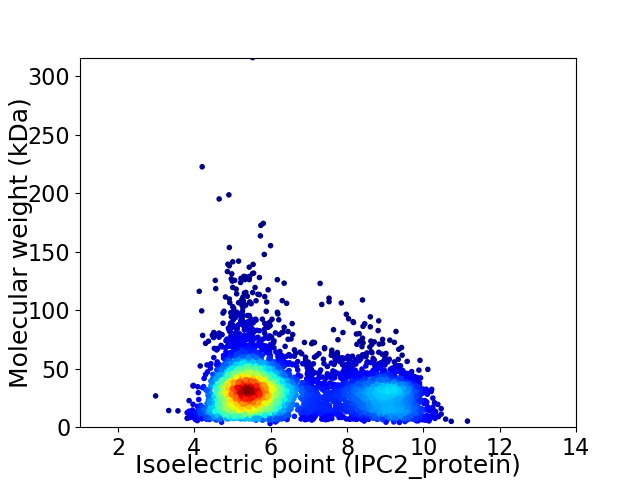
Virtual 2D-PAGE plot for 5159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8QN79|A0A4Y8QN79_9RHIZ Uncharacterized protein OS=Shinella sp. MEC087 OX=1967501 GN=B5M44_04845 PE=4 SV=1
MM1 pKa = 6.93VAAEE5 pKa = 4.66PEE7 pKa = 3.86PLEE10 pKa = 4.03YY11 pKa = 11.01VRR13 pKa = 11.84VCDD16 pKa = 4.21AFGTGFFYY24 pKa = 10.4IPGTEE29 pKa = 3.91TCLKK33 pKa = 9.25ISGYY37 pKa = 9.95VRR39 pKa = 11.84FQTDD43 pKa = 3.44FGRR46 pKa = 11.84DD47 pKa = 3.17EE48 pKa = 5.06FGNSDD53 pKa = 2.85WSSWTRR59 pKa = 11.84ARR61 pKa = 11.84VAFDD65 pKa = 3.56AKK67 pKa = 10.93SDD69 pKa = 4.02TEE71 pKa = 4.62LGTLTSFIAIHH82 pKa = 6.41GEE84 pKa = 4.09SGHH87 pKa = 6.52GGLGLDD93 pKa = 3.79IEE95 pKa = 4.39NAEE98 pKa = 4.12LVGLGSGASSDD109 pKa = 4.03FYY111 pKa = 10.9IDD113 pKa = 2.96EE114 pKa = 4.91AYY116 pKa = 9.83IEE118 pKa = 4.34LGGFRR123 pKa = 11.84VGTFLNWWDD132 pKa = 3.47NDD134 pKa = 4.48LIGEE138 pKa = 4.47TEE140 pKa = 4.01TASNITRR147 pKa = 11.84FTSVRR152 pKa = 11.84YY153 pKa = 10.0VYY155 pKa = 10.85DD156 pKa = 3.71GGTFRR161 pKa = 11.84AGLSVDD167 pKa = 4.96EE168 pKa = 4.96IPDD171 pKa = 3.64YY172 pKa = 11.45TDD174 pKa = 3.97LLTNATYY181 pKa = 10.8HH182 pKa = 6.14NGFGVSGMVAATLGGVNFEE201 pKa = 4.89LLGAYY206 pKa = 9.65DD207 pKa = 4.58DD208 pKa = 4.2STEE211 pKa = 4.06EE212 pKa = 4.03GAVRR216 pKa = 11.84AWLSAEE222 pKa = 3.98VGPGTFEE229 pKa = 5.42LMGLYY234 pKa = 10.49ASGPNAYY241 pKa = 9.82YY242 pKa = 10.6DD243 pKa = 3.51AAEE246 pKa = 4.04WSVAADD252 pKa = 3.51YY253 pKa = 11.22SFKK256 pKa = 10.55ATEE259 pKa = 4.59KK260 pKa = 10.32LTITPGAQYY269 pKa = 10.53FGDD272 pKa = 3.7YY273 pKa = 11.23GLIDD277 pKa = 5.1GNDD280 pKa = 3.18AWRR283 pKa = 11.84VGLTVDD289 pKa = 3.64YY290 pKa = 11.2AIVDD294 pKa = 3.84NLALKK299 pKa = 9.44ATVNYY304 pKa = 8.12TDD306 pKa = 5.2PDD308 pKa = 3.85NEE310 pKa = 4.64DD311 pKa = 3.81DD312 pKa = 4.32FVTGFLRR319 pKa = 11.84LQRR322 pKa = 11.84NFF324 pKa = 3.24
MM1 pKa = 6.93VAAEE5 pKa = 4.66PEE7 pKa = 3.86PLEE10 pKa = 4.03YY11 pKa = 11.01VRR13 pKa = 11.84VCDD16 pKa = 4.21AFGTGFFYY24 pKa = 10.4IPGTEE29 pKa = 3.91TCLKK33 pKa = 9.25ISGYY37 pKa = 9.95VRR39 pKa = 11.84FQTDD43 pKa = 3.44FGRR46 pKa = 11.84DD47 pKa = 3.17EE48 pKa = 5.06FGNSDD53 pKa = 2.85WSSWTRR59 pKa = 11.84ARR61 pKa = 11.84VAFDD65 pKa = 3.56AKK67 pKa = 10.93SDD69 pKa = 4.02TEE71 pKa = 4.62LGTLTSFIAIHH82 pKa = 6.41GEE84 pKa = 4.09SGHH87 pKa = 6.52GGLGLDD93 pKa = 3.79IEE95 pKa = 4.39NAEE98 pKa = 4.12LVGLGSGASSDD109 pKa = 4.03FYY111 pKa = 10.9IDD113 pKa = 2.96EE114 pKa = 4.91AYY116 pKa = 9.83IEE118 pKa = 4.34LGGFRR123 pKa = 11.84VGTFLNWWDD132 pKa = 3.47NDD134 pKa = 4.48LIGEE138 pKa = 4.47TEE140 pKa = 4.01TASNITRR147 pKa = 11.84FTSVRR152 pKa = 11.84YY153 pKa = 10.0VYY155 pKa = 10.85DD156 pKa = 3.71GGTFRR161 pKa = 11.84AGLSVDD167 pKa = 4.96EE168 pKa = 4.96IPDD171 pKa = 3.64YY172 pKa = 11.45TDD174 pKa = 3.97LLTNATYY181 pKa = 10.8HH182 pKa = 6.14NGFGVSGMVAATLGGVNFEE201 pKa = 4.89LLGAYY206 pKa = 9.65DD207 pKa = 4.58DD208 pKa = 4.2STEE211 pKa = 4.06EE212 pKa = 4.03GAVRR216 pKa = 11.84AWLSAEE222 pKa = 3.98VGPGTFEE229 pKa = 5.42LMGLYY234 pKa = 10.49ASGPNAYY241 pKa = 9.82YY242 pKa = 10.6DD243 pKa = 3.51AAEE246 pKa = 4.04WSVAADD252 pKa = 3.51YY253 pKa = 11.22SFKK256 pKa = 10.55ATEE259 pKa = 4.59KK260 pKa = 10.32LTITPGAQYY269 pKa = 10.53FGDD272 pKa = 3.7YY273 pKa = 11.23GLIDD277 pKa = 5.1GNDD280 pKa = 3.18AWRR283 pKa = 11.84VGLTVDD289 pKa = 3.64YY290 pKa = 11.2AIVDD294 pKa = 3.84NLALKK299 pKa = 9.44ATVNYY304 pKa = 8.12TDD306 pKa = 5.2PDD308 pKa = 3.85NEE310 pKa = 4.64DD311 pKa = 3.81DD312 pKa = 4.32FVTGFLRR319 pKa = 11.84LQRR322 pKa = 11.84NFF324 pKa = 3.24
Molecular weight: 35.27 kDa
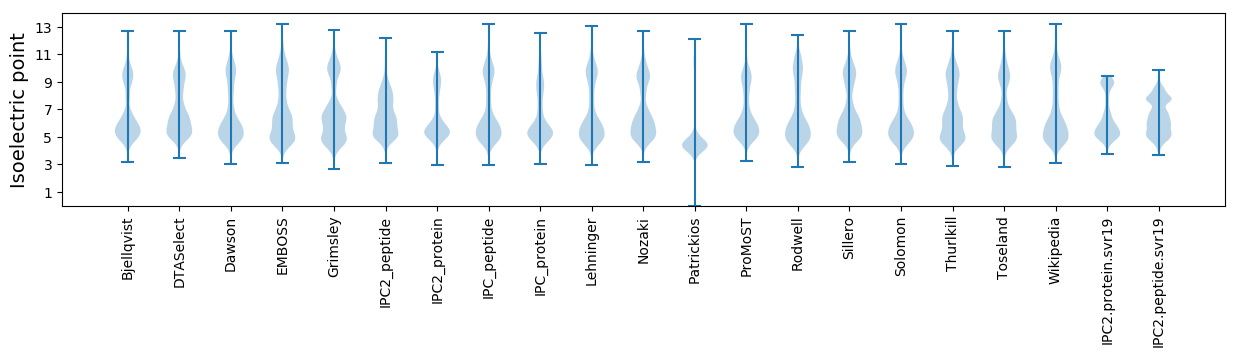
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8QMR8|A0A4Y8QMR8_9RHIZ RpiR family transcriptional regulator OS=Shinella sp. MEC087 OX=1967501 GN=B5M44_05715 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.27QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.22GGRR29 pKa = 11.84KK30 pKa = 9.06VLSARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.99
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.27QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.22GGRR29 pKa = 11.84KK30 pKa = 9.06VLSARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.99
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1578796 |
28 |
2833 |
306.0 |
33.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.467 ± 0.045 | 0.798 ± 0.01 |
5.764 ± 0.029 | 5.766 ± 0.031 |
3.853 ± 0.023 | 8.535 ± 0.033 |
1.986 ± 0.016 | 5.571 ± 0.022 |
3.573 ± 0.025 | 9.887 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.017 | 2.726 ± 0.02 |
4.801 ± 0.026 | 2.933 ± 0.021 |
6.827 ± 0.032 | 5.473 ± 0.026 |
5.433 ± 0.021 | 7.503 ± 0.027 |
1.257 ± 0.014 | 2.293 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
