
Botrimarina hoheduenensis
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Botrimarina
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
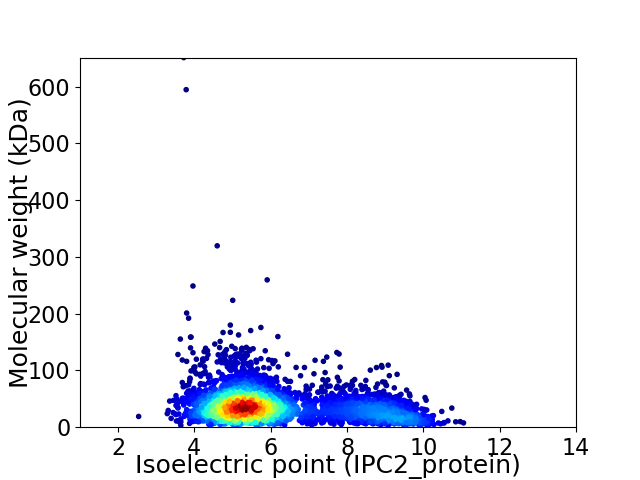
Virtual 2D-PAGE plot for 3424 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
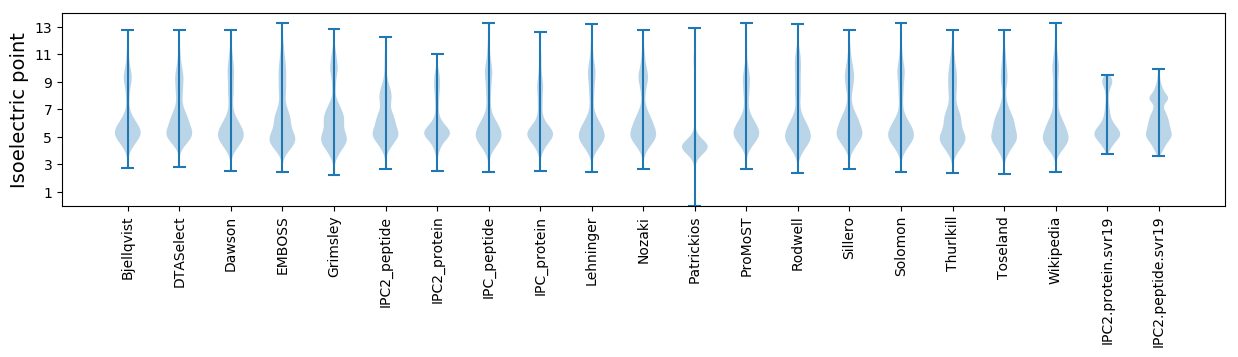
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5W9S5|A0A5C5W9S5_9BACT PhoD-like phosphatase OS=Botrimarina hoheduenensis OX=2528000 GN=Pla111_12440 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 8.05PTRR5 pKa = 11.84TRR7 pKa = 11.84SAHH10 pKa = 5.15YY11 pKa = 9.86SSPRR15 pKa = 11.84GGGRR19 pKa = 11.84FACLLTRR26 pKa = 11.84TAGLLLAAASVGEE39 pKa = 4.21VTGLEE44 pKa = 4.11IVVPAYY50 pKa = 9.47FYY52 pKa = 10.34PSSSGSDD59 pKa = 2.99WDD61 pKa = 4.49RR62 pKa = 11.84LDD64 pKa = 3.3TAAAAVPITAIMNPFNGPGNSFNSDD89 pKa = 3.07YY90 pKa = 11.59GSATNSFTSAGGNLIGYY107 pKa = 8.25VYY109 pKa = 9.86SQYY112 pKa = 11.12GARR115 pKa = 11.84PLNVVLADD123 pKa = 3.35IDD125 pKa = 5.88RR126 pKa = 11.84YY127 pKa = 8.77AQWYY131 pKa = 7.61PVDD134 pKa = 4.78GIFVDD139 pKa = 4.26EE140 pKa = 5.19FSNSSDD146 pKa = 3.54PAVLDD151 pKa = 3.85YY152 pKa = 11.71YY153 pKa = 11.04NAIYY157 pKa = 10.21QYY159 pKa = 11.09VKK161 pKa = 10.66SIDD164 pKa = 3.85PAWEE168 pKa = 3.99VMGNPGTTTVEE179 pKa = 4.04NYY181 pKa = 7.37LTRR184 pKa = 11.84PAADD188 pKa = 3.82RR189 pKa = 11.84LMIWEE194 pKa = 4.52NFGATYY200 pKa = 7.8PTQSPPSWTANYY212 pKa = 8.15DD213 pKa = 3.0ASRR216 pKa = 11.84FVHH219 pKa = 6.75LLHH222 pKa = 6.55TLGSSSTATDD232 pKa = 3.8YY233 pKa = 11.84VDD235 pKa = 4.01LAVARR240 pKa = 11.84NVGGVYY246 pKa = 8.65FTDD249 pKa = 5.26DD250 pKa = 4.03EE251 pKa = 5.41LPNPWDD257 pKa = 3.83RR258 pKa = 11.84LPTYY262 pKa = 9.94WDD264 pKa = 3.12AFVAKK269 pKa = 10.26VEE271 pKa = 4.45SVNNQVFGSLEE282 pKa = 3.87QLSNPVAEE290 pKa = 4.37GAIMIDD296 pKa = 3.55SSRR299 pKa = 11.84SDD301 pKa = 2.98WSGLSAYY308 pKa = 10.64SPDD311 pKa = 3.57VDD313 pKa = 4.07DD314 pKa = 4.43TTSSSLDD321 pKa = 3.26IVEE324 pKa = 4.2VTLANDD330 pKa = 3.63PDD332 pKa = 3.91EE333 pKa = 5.12LFVRR337 pKa = 11.84LTLDD341 pKa = 3.68GSTTPLGNGHH351 pKa = 7.09RR352 pKa = 11.84LLIDD356 pKa = 3.09VDD358 pKa = 4.01GQRR361 pKa = 11.84GTGYY365 pKa = 10.82LGGSEE370 pKa = 4.04QFALGADD377 pKa = 3.74YY378 pKa = 11.14LILSDD383 pKa = 5.18RR384 pKa = 11.84LFAFQGLTQDD394 pKa = 3.41VFSWSFLADD403 pKa = 4.26LSTDD407 pKa = 3.36DD408 pKa = 4.52STPSDD413 pKa = 3.87IEE415 pKa = 4.0WAILLSDD422 pKa = 4.57LGSPDD427 pKa = 3.18VLNFFVEE434 pKa = 4.96TTSSAGNDD442 pKa = 3.75YY443 pKa = 10.97LPSRR447 pKa = 11.84AVDD450 pKa = 3.62GLAGGFYY457 pKa = 9.69RR458 pKa = 11.84YY459 pKa = 10.09KK460 pKa = 10.46IGTPPVAGDD469 pKa = 3.46YY470 pKa = 11.34DD471 pKa = 4.33GDD473 pKa = 4.23GDD475 pKa = 4.78VDD477 pKa = 3.63TDD479 pKa = 5.05DD480 pKa = 4.48YY481 pKa = 11.56EE482 pKa = 4.11VWKK485 pKa = 11.09ANFGGPNLAADD496 pKa = 4.28GNDD499 pKa = 3.94DD500 pKa = 4.19GVIDD504 pKa = 4.04AADD507 pKa = 3.47YY508 pKa = 8.3TVWRR512 pKa = 11.84DD513 pKa = 3.08AAMLAGGATIPEE525 pKa = 4.42PSLGSLLVAILTAITVAPHH544 pKa = 6.35RR545 pKa = 11.84RR546 pKa = 11.84QEE548 pKa = 3.76
MM1 pKa = 7.56KK2 pKa = 8.05PTRR5 pKa = 11.84TRR7 pKa = 11.84SAHH10 pKa = 5.15YY11 pKa = 9.86SSPRR15 pKa = 11.84GGGRR19 pKa = 11.84FACLLTRR26 pKa = 11.84TAGLLLAAASVGEE39 pKa = 4.21VTGLEE44 pKa = 4.11IVVPAYY50 pKa = 9.47FYY52 pKa = 10.34PSSSGSDD59 pKa = 2.99WDD61 pKa = 4.49RR62 pKa = 11.84LDD64 pKa = 3.3TAAAAVPITAIMNPFNGPGNSFNSDD89 pKa = 3.07YY90 pKa = 11.59GSATNSFTSAGGNLIGYY107 pKa = 8.25VYY109 pKa = 9.86SQYY112 pKa = 11.12GARR115 pKa = 11.84PLNVVLADD123 pKa = 3.35IDD125 pKa = 5.88RR126 pKa = 11.84YY127 pKa = 8.77AQWYY131 pKa = 7.61PVDD134 pKa = 4.78GIFVDD139 pKa = 4.26EE140 pKa = 5.19FSNSSDD146 pKa = 3.54PAVLDD151 pKa = 3.85YY152 pKa = 11.71YY153 pKa = 11.04NAIYY157 pKa = 10.21QYY159 pKa = 11.09VKK161 pKa = 10.66SIDD164 pKa = 3.85PAWEE168 pKa = 3.99VMGNPGTTTVEE179 pKa = 4.04NYY181 pKa = 7.37LTRR184 pKa = 11.84PAADD188 pKa = 3.82RR189 pKa = 11.84LMIWEE194 pKa = 4.52NFGATYY200 pKa = 7.8PTQSPPSWTANYY212 pKa = 8.15DD213 pKa = 3.0ASRR216 pKa = 11.84FVHH219 pKa = 6.75LLHH222 pKa = 6.55TLGSSSTATDD232 pKa = 3.8YY233 pKa = 11.84VDD235 pKa = 4.01LAVARR240 pKa = 11.84NVGGVYY246 pKa = 8.65FTDD249 pKa = 5.26DD250 pKa = 4.03EE251 pKa = 5.41LPNPWDD257 pKa = 3.83RR258 pKa = 11.84LPTYY262 pKa = 9.94WDD264 pKa = 3.12AFVAKK269 pKa = 10.26VEE271 pKa = 4.45SVNNQVFGSLEE282 pKa = 3.87QLSNPVAEE290 pKa = 4.37GAIMIDD296 pKa = 3.55SSRR299 pKa = 11.84SDD301 pKa = 2.98WSGLSAYY308 pKa = 10.64SPDD311 pKa = 3.57VDD313 pKa = 4.07DD314 pKa = 4.43TTSSSLDD321 pKa = 3.26IVEE324 pKa = 4.2VTLANDD330 pKa = 3.63PDD332 pKa = 3.91EE333 pKa = 5.12LFVRR337 pKa = 11.84LTLDD341 pKa = 3.68GSTTPLGNGHH351 pKa = 7.09RR352 pKa = 11.84LLIDD356 pKa = 3.09VDD358 pKa = 4.01GQRR361 pKa = 11.84GTGYY365 pKa = 10.82LGGSEE370 pKa = 4.04QFALGADD377 pKa = 3.74YY378 pKa = 11.14LILSDD383 pKa = 5.18RR384 pKa = 11.84LFAFQGLTQDD394 pKa = 3.41VFSWSFLADD403 pKa = 4.26LSTDD407 pKa = 3.36DD408 pKa = 4.52STPSDD413 pKa = 3.87IEE415 pKa = 4.0WAILLSDD422 pKa = 4.57LGSPDD427 pKa = 3.18VLNFFVEE434 pKa = 4.96TTSSAGNDD442 pKa = 3.75YY443 pKa = 10.97LPSRR447 pKa = 11.84AVDD450 pKa = 3.62GLAGGFYY457 pKa = 9.69RR458 pKa = 11.84YY459 pKa = 10.09KK460 pKa = 10.46IGTPPVAGDD469 pKa = 3.46YY470 pKa = 11.34DD471 pKa = 4.33GDD473 pKa = 4.23GDD475 pKa = 4.78VDD477 pKa = 3.63TDD479 pKa = 5.05DD480 pKa = 4.48YY481 pKa = 11.56EE482 pKa = 4.11VWKK485 pKa = 11.09ANFGGPNLAADD496 pKa = 4.28GNDD499 pKa = 3.94DD500 pKa = 4.19GVIDD504 pKa = 4.04AADD507 pKa = 3.47YY508 pKa = 8.3TVWRR512 pKa = 11.84DD513 pKa = 3.08AAMLAGGATIPEE525 pKa = 4.42PSLGSLLVAILTAITVAPHH544 pKa = 6.35RR545 pKa = 11.84RR546 pKa = 11.84QEE548 pKa = 3.76
Molecular weight: 58.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5VTS3|A0A5C5VTS3_9BACT Mercuric reductase OS=Botrimarina hoheduenensis OX=2528000 GN=merA PE=3 SV=1
MM1 pKa = 7.57TKK3 pKa = 10.2PSSPGLNKK11 pKa = 9.47TLVMTRR17 pKa = 11.84LLVAKK22 pKa = 10.02FGMAAGEE29 pKa = 3.94PVRR32 pKa = 11.84TKK34 pKa = 10.25KK35 pKa = 10.05KK36 pKa = 10.27RR37 pKa = 11.84RR38 pKa = 11.84IAPKK42 pKa = 8.09PKK44 pKa = 8.94RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84PKK52 pKa = 9.35PQLKK56 pKa = 9.63RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84APRR62 pKa = 11.84PKK64 pKa = 10.18RR65 pKa = 3.49
MM1 pKa = 7.57TKK3 pKa = 10.2PSSPGLNKK11 pKa = 9.47TLVMTRR17 pKa = 11.84LLVAKK22 pKa = 10.02FGMAAGEE29 pKa = 3.94PVRR32 pKa = 11.84TKK34 pKa = 10.25KK35 pKa = 10.05KK36 pKa = 10.27RR37 pKa = 11.84RR38 pKa = 11.84IAPKK42 pKa = 8.09PKK44 pKa = 8.94RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84PKK52 pKa = 9.35PQLKK56 pKa = 9.63RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84APRR62 pKa = 11.84PKK64 pKa = 10.18RR65 pKa = 3.49
Molecular weight: 7.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1258017 |
30 |
6319 |
367.4 |
39.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.926 ± 0.056 | 1.048 ± 0.02 |
5.882 ± 0.043 | 6.128 ± 0.044 |
3.323 ± 0.024 | 8.441 ± 0.054 |
1.938 ± 0.021 | 4.168 ± 0.023 |
2.71 ± 0.037 | 10.356 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.851 ± 0.021 | 2.754 ± 0.029 |
5.643 ± 0.038 | 3.601 ± 0.024 |
7.257 ± 0.046 | 5.924 ± 0.034 |
5.76 ± 0.039 | 7.456 ± 0.031 |
1.476 ± 0.02 | 2.357 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
