
Synechococcus sp. RSCCF101
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
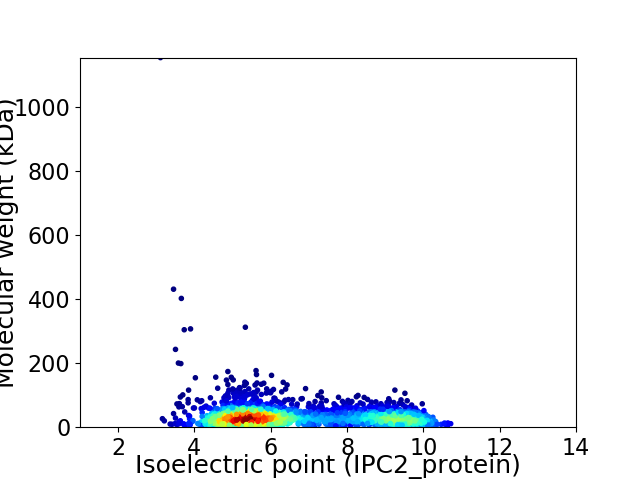
Virtual 2D-PAGE plot for 2660 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J6Q531|A0A5J6Q531_9SYNE Peptidyl-tRNA hydrolase OS=Synechococcus sp. RSCCF101 OX=2511069 GN=pth PE=3 SV=1
MM1 pKa = 6.81QWEE4 pKa = 4.41FQEE7 pKa = 4.8DD8 pKa = 3.69AAFLALCDD16 pKa = 3.64AFKK19 pKa = 11.03EE20 pKa = 4.38SGEE23 pKa = 4.06SSAIEE28 pKa = 3.79FLAHH32 pKa = 6.67GEE34 pKa = 4.38GAFHH38 pKa = 6.71FQEE41 pKa = 4.32LTQNAAGEE49 pKa = 4.49GVDD52 pKa = 5.68LSDD55 pKa = 5.64SDD57 pKa = 5.03SLDD60 pKa = 4.09DD61 pKa = 4.2FQQEE65 pKa = 4.46VIDD68 pKa = 4.26TMEE71 pKa = 4.54ALCSS75 pKa = 3.6
MM1 pKa = 6.81QWEE4 pKa = 4.41FQEE7 pKa = 4.8DD8 pKa = 3.69AAFLALCDD16 pKa = 3.64AFKK19 pKa = 11.03EE20 pKa = 4.38SGEE23 pKa = 4.06SSAIEE28 pKa = 3.79FLAHH32 pKa = 6.67GEE34 pKa = 4.38GAFHH38 pKa = 6.71FQEE41 pKa = 4.32LTQNAAGEE49 pKa = 4.49GVDD52 pKa = 5.68LSDD55 pKa = 5.64SDD57 pKa = 5.03SLDD60 pKa = 4.09DD61 pKa = 4.2FQQEE65 pKa = 4.46VIDD68 pKa = 4.26TMEE71 pKa = 4.54ALCSS75 pKa = 3.6
Molecular weight: 8.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J6PY12|A0A5J6PY12_9SYNE Site-specific integrase OS=Synechococcus sp. RSCCF101 OX=2511069 GN=EVJ50_02990 PE=4 SV=1
MM1 pKa = 7.51GSDD4 pKa = 3.24HH5 pKa = 6.97HH6 pKa = 8.08RR7 pKa = 11.84GEE9 pKa = 4.37PSVGTEE15 pKa = 3.54TWRR18 pKa = 11.84QRR20 pKa = 11.84TLLRR24 pKa = 11.84DD25 pKa = 4.04FDD27 pKa = 3.73RR28 pKa = 11.84TAWLLRR34 pKa = 11.84ADD36 pKa = 4.12LVQRR40 pKa = 11.84YY41 pKa = 7.78GEE43 pKa = 4.02RR44 pKa = 11.84DD45 pKa = 3.21AEE47 pKa = 3.92RR48 pKa = 11.84LARR51 pKa = 11.84HH52 pKa = 5.2ARR54 pKa = 11.84VRR56 pKa = 11.84CMEE59 pKa = 3.98ILGSVSWVKK68 pKa = 10.73GRR70 pKa = 11.84GAAVMNGFLWISAQEE85 pKa = 3.89LALFQAIEE93 pKa = 3.98ARR95 pKa = 11.84GGDD98 pKa = 3.78AAEE101 pKa = 4.25AWAICHH107 pKa = 5.7KK108 pKa = 10.48AIRR111 pKa = 11.84LRR113 pKa = 11.84VEE115 pKa = 5.62RR116 pKa = 11.84IPGWKK121 pKa = 8.48RR122 pKa = 11.84WLMRR126 pKa = 11.84RR127 pKa = 11.84LLFSALVRR135 pKa = 11.84RR136 pKa = 11.84IIRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84AVRR144 pKa = 11.84GDD146 pKa = 3.11IVRR149 pKa = 11.84AGDD152 pKa = 3.58FKK154 pKa = 10.81TRR156 pKa = 11.84SLIGDD161 pKa = 4.63GIAFDD166 pKa = 5.02LGVDD170 pKa = 3.56YY171 pKa = 10.79LRR173 pKa = 11.84CGNLEE178 pKa = 3.94LARR181 pKa = 11.84TIGAEE186 pKa = 3.77TFAPYY191 pKa = 10.74LCLSDD196 pKa = 3.6IALSEE201 pKa = 4.01GLGWGLIRR209 pKa = 11.84TQTLADD215 pKa = 4.01GCSHH219 pKa = 7.09CDD221 pKa = 3.51FRR223 pKa = 11.84FKK225 pKa = 10.81QGGPTRR231 pKa = 11.84IKK233 pKa = 10.54SRR235 pKa = 11.84TPEE238 pKa = 3.76VQATLEE244 pKa = 4.35RR245 pKa = 11.84IARR248 pKa = 11.84TEE250 pKa = 4.01AEE252 pKa = 4.25LAATAQSLRR261 pKa = 3.77
MM1 pKa = 7.51GSDD4 pKa = 3.24HH5 pKa = 6.97HH6 pKa = 8.08RR7 pKa = 11.84GEE9 pKa = 4.37PSVGTEE15 pKa = 3.54TWRR18 pKa = 11.84QRR20 pKa = 11.84TLLRR24 pKa = 11.84DD25 pKa = 4.04FDD27 pKa = 3.73RR28 pKa = 11.84TAWLLRR34 pKa = 11.84ADD36 pKa = 4.12LVQRR40 pKa = 11.84YY41 pKa = 7.78GEE43 pKa = 4.02RR44 pKa = 11.84DD45 pKa = 3.21AEE47 pKa = 3.92RR48 pKa = 11.84LARR51 pKa = 11.84HH52 pKa = 5.2ARR54 pKa = 11.84VRR56 pKa = 11.84CMEE59 pKa = 3.98ILGSVSWVKK68 pKa = 10.73GRR70 pKa = 11.84GAAVMNGFLWISAQEE85 pKa = 3.89LALFQAIEE93 pKa = 3.98ARR95 pKa = 11.84GGDD98 pKa = 3.78AAEE101 pKa = 4.25AWAICHH107 pKa = 5.7KK108 pKa = 10.48AIRR111 pKa = 11.84LRR113 pKa = 11.84VEE115 pKa = 5.62RR116 pKa = 11.84IPGWKK121 pKa = 8.48RR122 pKa = 11.84WLMRR126 pKa = 11.84RR127 pKa = 11.84LLFSALVRR135 pKa = 11.84RR136 pKa = 11.84IIRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84AVRR144 pKa = 11.84GDD146 pKa = 3.11IVRR149 pKa = 11.84AGDD152 pKa = 3.58FKK154 pKa = 10.81TRR156 pKa = 11.84SLIGDD161 pKa = 4.63GIAFDD166 pKa = 5.02LGVDD170 pKa = 3.56YY171 pKa = 10.79LRR173 pKa = 11.84CGNLEE178 pKa = 3.94LARR181 pKa = 11.84TIGAEE186 pKa = 3.77TFAPYY191 pKa = 10.74LCLSDD196 pKa = 3.6IALSEE201 pKa = 4.01GLGWGLIRR209 pKa = 11.84TQTLADD215 pKa = 4.01GCSHH219 pKa = 7.09CDD221 pKa = 3.51FRR223 pKa = 11.84FKK225 pKa = 10.81QGGPTRR231 pKa = 11.84IKK233 pKa = 10.54SRR235 pKa = 11.84TPEE238 pKa = 3.76VQATLEE244 pKa = 4.35RR245 pKa = 11.84IARR248 pKa = 11.84TEE250 pKa = 4.01AEE252 pKa = 4.25LAATAQSLRR261 pKa = 3.77
Molecular weight: 29.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
856344 |
30 |
11376 |
321.9 |
34.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.741 ± 0.063 | 1.163 ± 0.025 |
5.452 ± 0.115 | 5.849 ± 0.052 |
2.851 ± 0.039 | 8.963 ± 0.065 |
2.077 ± 0.033 | 3.779 ± 0.041 |
1.613 ± 0.029 | 12.578 ± 0.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.831 ± 0.033 | 2.06 ± 0.042 |
6.208 ± 0.056 | 4.003 ± 0.04 |
8.457 ± 0.115 | 6.377 ± 0.075 |
4.761 ± 0.101 | 6.894 ± 0.05 |
1.695 ± 0.031 | 1.649 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
