
Soft-shelled turtle iridovirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Iridoviridae; Alphairidovirinae; Ranavirus; Frog virus 3
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
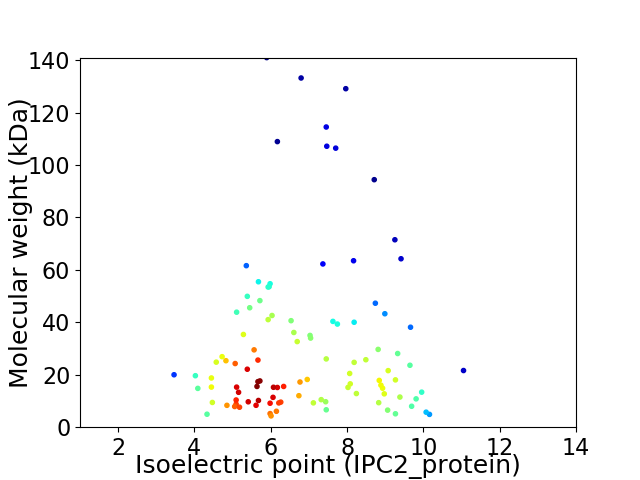
Virtual 2D-PAGE plot for 105 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
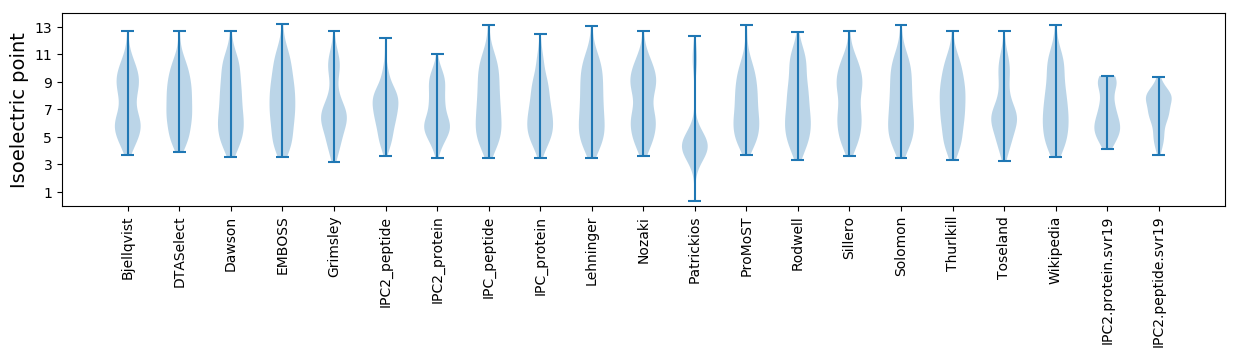
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C3RWT1|C3RWT1_FRG3V Putative membrane associated motif in LPS-induced tumor necrosis factor alpha OS=Soft-shelled turtle iridovirus OX=365144 GN=ORF082L PE=4 SV=1
MM1 pKa = 7.14VLLMSSVGLRR11 pKa = 11.84CLAALPYY18 pKa = 8.55TWRR21 pKa = 11.84LPGVICDD28 pKa = 3.63PWTGPTGTLDD38 pKa = 3.65SEE40 pKa = 4.99CPVATGSGALKK51 pKa = 10.64ASLCLEE57 pKa = 4.2YY58 pKa = 10.87AFSIPAGAGGGSLWAPDD75 pKa = 3.87PMPWTPLAVPPAMMLSAVVRR95 pKa = 11.84GARR98 pKa = 11.84LTALEE103 pKa = 4.12NWSLNPAWPGVWGLGVGSLRR123 pKa = 11.84TADD126 pKa = 4.71PPFPWEE132 pKa = 4.12PTSSMMGEE140 pKa = 4.11EE141 pKa = 4.5DD142 pKa = 4.43CLCGAA147 pKa = 5.24
MM1 pKa = 7.14VLLMSSVGLRR11 pKa = 11.84CLAALPYY18 pKa = 8.55TWRR21 pKa = 11.84LPGVICDD28 pKa = 3.63PWTGPTGTLDD38 pKa = 3.65SEE40 pKa = 4.99CPVATGSGALKK51 pKa = 10.64ASLCLEE57 pKa = 4.2YY58 pKa = 10.87AFSIPAGAGGGSLWAPDD75 pKa = 3.87PMPWTPLAVPPAMMLSAVVRR95 pKa = 11.84GARR98 pKa = 11.84LTALEE103 pKa = 4.12NWSLNPAWPGVWGLGVGSLRR123 pKa = 11.84TADD126 pKa = 4.71PPFPWEE132 pKa = 4.12PTSSMMGEE140 pKa = 4.11EE141 pKa = 4.5DD142 pKa = 4.43CLCGAA147 pKa = 5.24
Molecular weight: 15.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C3RWP9|C3RWP9_FRG3V Uncharacterized protein OS=Soft-shelled turtle iridovirus OX=365144 GN=ORF050L PE=4 SV=1
MM1 pKa = 7.11YY2 pKa = 10.24SVRR5 pKa = 11.84NSGCSAGCSPRR16 pKa = 11.84QGASPIMFGPSLGAMLSAPVVRR38 pKa = 11.84ASSPVVKK45 pKa = 10.32RR46 pKa = 11.84KK47 pKa = 10.35SLVKK51 pKa = 10.26RR52 pKa = 11.84KK53 pKa = 10.2SPVKK57 pKa = 10.34RR58 pKa = 11.84SPLKK62 pKa = 10.44KK63 pKa = 9.82RR64 pKa = 11.84SQTRR68 pKa = 11.84TSPVKK73 pKa = 10.24RR74 pKa = 11.84KK75 pKa = 10.42SPVKK79 pKa = 10.07RR80 pKa = 11.84KK81 pKa = 10.4SPVKK85 pKa = 10.07RR86 pKa = 11.84KK87 pKa = 10.4SPVKK91 pKa = 10.07RR92 pKa = 11.84KK93 pKa = 10.4SPVKK97 pKa = 10.07RR98 pKa = 11.84KK99 pKa = 10.4SPVKK103 pKa = 10.07RR104 pKa = 11.84KK105 pKa = 10.4SPVKK109 pKa = 10.07RR110 pKa = 11.84KK111 pKa = 10.4SPVKK115 pKa = 10.07RR116 pKa = 11.84KK117 pKa = 10.4SPVKK121 pKa = 10.07RR122 pKa = 11.84KK123 pKa = 10.4SPVKK127 pKa = 10.07RR128 pKa = 11.84KK129 pKa = 10.4SPVKK133 pKa = 10.07RR134 pKa = 11.84KK135 pKa = 10.4SPVKK139 pKa = 10.07RR140 pKa = 11.84KK141 pKa = 10.4SPVKK145 pKa = 10.07RR146 pKa = 11.84KK147 pKa = 10.4SPVKK151 pKa = 10.07RR152 pKa = 11.84KK153 pKa = 10.4SPVKK157 pKa = 10.07RR158 pKa = 11.84KK159 pKa = 10.4SPVKK163 pKa = 10.07RR164 pKa = 11.84KK165 pKa = 10.4SPVKK169 pKa = 10.07RR170 pKa = 11.84KK171 pKa = 10.4SPVKK175 pKa = 10.05RR176 pKa = 11.84KK177 pKa = 10.07SPAKK181 pKa = 10.0RR182 pKa = 11.84RR183 pKa = 11.84QQVCFMATKK192 pKa = 10.43
MM1 pKa = 7.11YY2 pKa = 10.24SVRR5 pKa = 11.84NSGCSAGCSPRR16 pKa = 11.84QGASPIMFGPSLGAMLSAPVVRR38 pKa = 11.84ASSPVVKK45 pKa = 10.32RR46 pKa = 11.84KK47 pKa = 10.35SLVKK51 pKa = 10.26RR52 pKa = 11.84KK53 pKa = 10.2SPVKK57 pKa = 10.34RR58 pKa = 11.84SPLKK62 pKa = 10.44KK63 pKa = 9.82RR64 pKa = 11.84SQTRR68 pKa = 11.84TSPVKK73 pKa = 10.24RR74 pKa = 11.84KK75 pKa = 10.42SPVKK79 pKa = 10.07RR80 pKa = 11.84KK81 pKa = 10.4SPVKK85 pKa = 10.07RR86 pKa = 11.84KK87 pKa = 10.4SPVKK91 pKa = 10.07RR92 pKa = 11.84KK93 pKa = 10.4SPVKK97 pKa = 10.07RR98 pKa = 11.84KK99 pKa = 10.4SPVKK103 pKa = 10.07RR104 pKa = 11.84KK105 pKa = 10.4SPVKK109 pKa = 10.07RR110 pKa = 11.84KK111 pKa = 10.4SPVKK115 pKa = 10.07RR116 pKa = 11.84KK117 pKa = 10.4SPVKK121 pKa = 10.07RR122 pKa = 11.84KK123 pKa = 10.4SPVKK127 pKa = 10.07RR128 pKa = 11.84KK129 pKa = 10.4SPVKK133 pKa = 10.07RR134 pKa = 11.84KK135 pKa = 10.4SPVKK139 pKa = 10.07RR140 pKa = 11.84KK141 pKa = 10.4SPVKK145 pKa = 10.07RR146 pKa = 11.84KK147 pKa = 10.4SPVKK151 pKa = 10.07RR152 pKa = 11.84KK153 pKa = 10.4SPVKK157 pKa = 10.07RR158 pKa = 11.84KK159 pKa = 10.4SPVKK163 pKa = 10.07RR164 pKa = 11.84KK165 pKa = 10.4SPVKK169 pKa = 10.07RR170 pKa = 11.84KK171 pKa = 10.4SPVKK175 pKa = 10.05RR176 pKa = 11.84KK177 pKa = 10.07SPAKK181 pKa = 10.0RR182 pKa = 11.84RR183 pKa = 11.84QQVCFMATKK192 pKa = 10.43
Molecular weight: 21.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
29450 |
42 |
1294 |
280.5 |
30.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.015 ± 0.275 | 2.105 ± 0.164 |
6.068 ± 0.152 | 5.657 ± 0.205 |
3.345 ± 0.115 | 7.324 ± 0.266 |
1.922 ± 0.128 | 3.284 ± 0.133 |
5.701 ± 0.309 | 8.143 ± 0.326 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.127 | 2.608 ± 0.125 |
6.278 ± 0.308 | 2.635 ± 0.149 |
6.788 ± 0.3 | 7.535 ± 0.246 |
5.671 ± 0.179 | 8.323 ± 0.222 |
1.501 ± 0.109 | 2.924 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
