
Sparrow coronavirus HKU17
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Deltacoronavirus; Buldecovirus; unclassified Buldecovirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
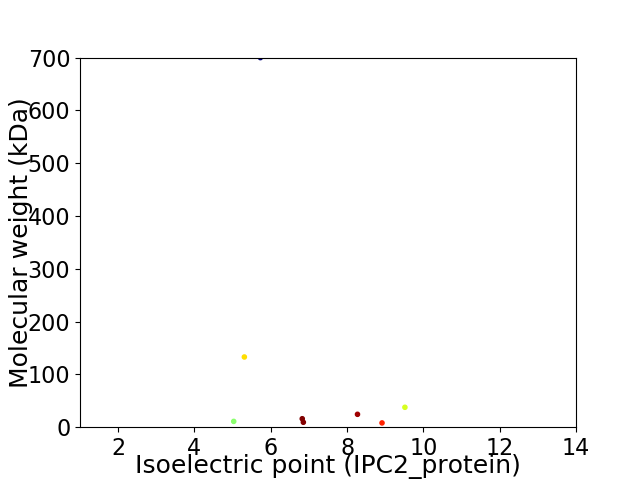
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9BR04|H9BR04_9NIDO Nucleocapsid protein OS=Sparrow coronavirus HKU17 OX=1159906 GN=N PE=4 SV=1
MM1 pKa = 7.59CNCLLQLRR9 pKa = 11.84EE10 pKa = 4.25LYY12 pKa = 10.43KK13 pKa = 10.9LCNEE17 pKa = 4.26RR18 pKa = 11.84NITRR22 pKa = 11.84DD23 pKa = 3.51DD24 pKa = 3.7VLEE27 pKa = 5.34LIDD30 pKa = 4.45PLIKK34 pKa = 9.74TRR36 pKa = 11.84CFAYY40 pKa = 10.63SLVVLANANPIALSILPRR58 pKa = 11.84KK59 pKa = 9.53ILINGEE65 pKa = 4.19PLLLEE70 pKa = 4.08YY71 pKa = 11.25GNIYY75 pKa = 10.59GKK77 pKa = 10.45DD78 pKa = 3.43FLYY81 pKa = 10.53RR82 pKa = 11.84PSLQVILEE90 pKa = 4.12EE91 pKa = 4.63EE92 pKa = 4.27EE93 pKa = 4.51LNN95 pKa = 4.07
MM1 pKa = 7.59CNCLLQLRR9 pKa = 11.84EE10 pKa = 4.25LYY12 pKa = 10.43KK13 pKa = 10.9LCNEE17 pKa = 4.26RR18 pKa = 11.84NITRR22 pKa = 11.84DD23 pKa = 3.51DD24 pKa = 3.7VLEE27 pKa = 5.34LIDD30 pKa = 4.45PLIKK34 pKa = 9.74TRR36 pKa = 11.84CFAYY40 pKa = 10.63SLVVLANANPIALSILPRR58 pKa = 11.84KK59 pKa = 9.53ILINGEE65 pKa = 4.19PLLLEE70 pKa = 4.08YY71 pKa = 11.25GNIYY75 pKa = 10.59GKK77 pKa = 10.45DD78 pKa = 3.43FLYY81 pKa = 10.53RR82 pKa = 11.84PSLQVILEE90 pKa = 4.12EE91 pKa = 4.63EE92 pKa = 4.27EE93 pKa = 4.51LNN95 pKa = 4.07
Molecular weight: 11.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9BR05|H9BR05_9NIDO NS7a protein OS=Sparrow coronavirus HKU17 OX=1159906 GN=NS7a PE=4 SV=1
MM1 pKa = 7.44ATPSVPTTDD10 pKa = 4.06ASWFQVLKK18 pKa = 10.58AQNKK22 pKa = 9.03KK23 pKa = 8.6ATHH26 pKa = 5.19PQFRR30 pKa = 11.84GNGVPLNSAIKK41 pKa = 8.82PAEE44 pKa = 3.75NHH46 pKa = 6.51GYY48 pKa = 7.44WLRR51 pKa = 11.84YY52 pKa = 7.01TRR54 pKa = 11.84QKK56 pKa = 10.76PGGTQIPPSYY66 pKa = 10.58AFYY69 pKa = 9.95YY70 pKa = 9.86TGTGPRR76 pKa = 11.84GNLKK80 pKa = 10.6YY81 pKa = 11.01GEE83 pKa = 4.87LPPNDD88 pKa = 3.7TQATTRR94 pKa = 11.84VTWVKK99 pKa = 10.92GPGADD104 pKa = 3.37TSIKK108 pKa = 9.68PHH110 pKa = 4.45VAKK113 pKa = 10.68RR114 pKa = 11.84NPNNPKK120 pKa = 9.85HH121 pKa = 5.96QLLPLRR127 pKa = 11.84FPTGDD132 pKa = 3.2GPAQGFRR139 pKa = 11.84VDD141 pKa = 3.64PFNARR146 pKa = 11.84GRR148 pKa = 11.84PQEE151 pKa = 4.34RR152 pKa = 11.84GSGPRR157 pKa = 11.84SQSANPRR164 pKa = 11.84GTNNQPRR171 pKa = 11.84KK172 pKa = 9.28RR173 pKa = 11.84DD174 pKa = 3.4QSAPAAVRR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 7.09TQHH187 pKa = 5.18QAPKK191 pKa = 9.09RR192 pKa = 11.84TLPKK196 pKa = 10.35GKK198 pKa = 8.69TISQVFGNRR207 pKa = 11.84SRR209 pKa = 11.84TGANVGSADD218 pKa = 3.79TEE220 pKa = 4.29KK221 pKa = 10.47TGMADD226 pKa = 3.45PRR228 pKa = 11.84IMALARR234 pKa = 11.84HH235 pKa = 5.28VPGVQEE241 pKa = 3.98MLFAGHH247 pKa = 7.25LEE249 pKa = 4.47SNFQAGAITLTFSYY263 pKa = 10.44SITVKK268 pKa = 10.43EE269 pKa = 4.05GSPDD273 pKa = 3.52YY274 pKa = 11.08EE275 pKa = 3.99RR276 pKa = 11.84LKK278 pKa = 11.02DD279 pKa = 3.7ALNTVVNQTYY289 pKa = 9.4EE290 pKa = 4.24PPTKK294 pKa = 8.96PTKK297 pKa = 10.18DD298 pKa = 3.39KK299 pKa = 11.2KK300 pKa = 10.44PEE302 pKa = 4.08KK303 pKa = 9.57QDD305 pKa = 3.24QSAKK309 pKa = 10.18PKK311 pKa = 8.42QQKK314 pKa = 9.59KK315 pKa = 8.18PKK317 pKa = 9.94KK318 pKa = 8.31VTLPADD324 pKa = 3.79KK325 pKa = 10.5QDD327 pKa = 4.98LEE329 pKa = 4.12WDD331 pKa = 3.51DD332 pKa = 4.14AFEE335 pKa = 4.59IKK337 pKa = 10.2QEE339 pKa = 4.19SAAA342 pKa = 4.0
MM1 pKa = 7.44ATPSVPTTDD10 pKa = 4.06ASWFQVLKK18 pKa = 10.58AQNKK22 pKa = 9.03KK23 pKa = 8.6ATHH26 pKa = 5.19PQFRR30 pKa = 11.84GNGVPLNSAIKK41 pKa = 8.82PAEE44 pKa = 3.75NHH46 pKa = 6.51GYY48 pKa = 7.44WLRR51 pKa = 11.84YY52 pKa = 7.01TRR54 pKa = 11.84QKK56 pKa = 10.76PGGTQIPPSYY66 pKa = 10.58AFYY69 pKa = 9.95YY70 pKa = 9.86TGTGPRR76 pKa = 11.84GNLKK80 pKa = 10.6YY81 pKa = 11.01GEE83 pKa = 4.87LPPNDD88 pKa = 3.7TQATTRR94 pKa = 11.84VTWVKK99 pKa = 10.92GPGADD104 pKa = 3.37TSIKK108 pKa = 9.68PHH110 pKa = 4.45VAKK113 pKa = 10.68RR114 pKa = 11.84NPNNPKK120 pKa = 9.85HH121 pKa = 5.96QLLPLRR127 pKa = 11.84FPTGDD132 pKa = 3.2GPAQGFRR139 pKa = 11.84VDD141 pKa = 3.64PFNARR146 pKa = 11.84GRR148 pKa = 11.84PQEE151 pKa = 4.34RR152 pKa = 11.84GSGPRR157 pKa = 11.84SQSANPRR164 pKa = 11.84GTNNQPRR171 pKa = 11.84KK172 pKa = 9.28RR173 pKa = 11.84DD174 pKa = 3.4QSAPAAVRR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 7.09TQHH187 pKa = 5.18QAPKK191 pKa = 9.09RR192 pKa = 11.84TLPKK196 pKa = 10.35GKK198 pKa = 8.69TISQVFGNRR207 pKa = 11.84SRR209 pKa = 11.84TGANVGSADD218 pKa = 3.79TEE220 pKa = 4.29KK221 pKa = 10.47TGMADD226 pKa = 3.45PRR228 pKa = 11.84IMALARR234 pKa = 11.84HH235 pKa = 5.28VPGVQEE241 pKa = 3.98MLFAGHH247 pKa = 7.25LEE249 pKa = 4.47SNFQAGAITLTFSYY263 pKa = 10.44SITVKK268 pKa = 10.43EE269 pKa = 4.05GSPDD273 pKa = 3.52YY274 pKa = 11.08EE275 pKa = 3.99RR276 pKa = 11.84LKK278 pKa = 11.02DD279 pKa = 3.7ALNTVVNQTYY289 pKa = 9.4EE290 pKa = 4.24PPTKK294 pKa = 8.96PTKK297 pKa = 10.18DD298 pKa = 3.39KK299 pKa = 11.2KK300 pKa = 10.44PEE302 pKa = 4.08KK303 pKa = 9.57QDD305 pKa = 3.24QSAKK309 pKa = 10.18PKK311 pKa = 8.42QQKK314 pKa = 9.59KK315 pKa = 8.18PKK317 pKa = 9.94KK318 pKa = 8.31VTLPADD324 pKa = 3.79KK325 pKa = 10.5QDD327 pKa = 4.98LEE329 pKa = 4.12WDD331 pKa = 3.51DD332 pKa = 4.14AFEE335 pKa = 4.59IKK337 pKa = 10.2QEE339 pKa = 4.19SAAA342 pKa = 4.0
Molecular weight: 37.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8433 |
70 |
6277 |
1054.1 |
117.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.909 ± 0.449 | 2.561 ± 0.335 |
5.277 ± 0.592 | 4.162 ± 0.318 |
4.316 ± 0.317 | 5.182 ± 0.541 |
2.502 ± 0.29 | 5.977 ± 0.663 |
5.052 ± 0.622 | 9.321 ± 0.578 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.719 ± 0.248 | 5.443 ± 0.535 |
5.099 ± 0.9 | 4.328 ± 0.391 |
3.581 ± 0.463 | 6.143 ± 0.628 |
7.862 ± 0.795 | 7.791 ± 0.881 |
0.901 ± 0.384 | 4.838 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
