
Hubei tombus-like virus 14
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
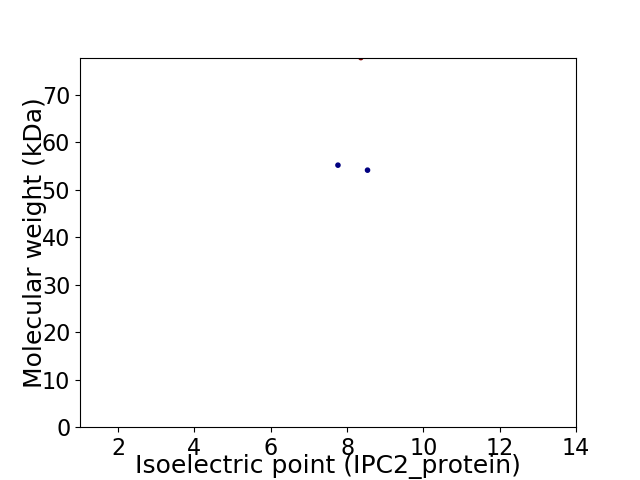
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGE5|A0A1L3KGE5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 14 OX=1923260 PE=4 SV=1
MM1 pKa = 7.31KK2 pKa = 10.21TPDD5 pKa = 3.67PQVNSAEE12 pKa = 3.98WKK14 pKa = 9.38QFRR17 pKa = 11.84KK18 pKa = 9.9VLRR21 pKa = 11.84EE22 pKa = 3.77LSDD25 pKa = 3.59EE26 pKa = 4.28VGSVPKK32 pKa = 9.55ATPTTVVNHH41 pKa = 6.13RR42 pKa = 11.84NSMKK46 pKa = 10.06KK47 pKa = 9.61RR48 pKa = 11.84RR49 pKa = 11.84FGKK52 pKa = 10.44GMEE55 pKa = 3.76IYY57 pKa = 10.52LRR59 pKa = 11.84EE60 pKa = 4.01GVAAKK65 pKa = 10.22HH66 pKa = 6.27SYY68 pKa = 7.71ITMMPKK74 pKa = 10.38LEE76 pKa = 4.87FYY78 pKa = 10.72DD79 pKa = 3.99SEE81 pKa = 4.62KK82 pKa = 10.74IPLKK86 pKa = 9.87EE87 pKa = 4.25DD88 pKa = 2.72RR89 pKa = 11.84GIQYY93 pKa = 10.04RR94 pKa = 11.84SPVYY98 pKa = 9.31NAALARR104 pKa = 11.84HH105 pKa = 5.67LHH107 pKa = 5.59HH108 pKa = 8.33VEE110 pKa = 3.6EE111 pKa = 4.28QLYY114 pKa = 8.12KK115 pKa = 10.18TIKK118 pKa = 10.47NVDD121 pKa = 3.59GTPVFAKK128 pKa = 10.05GYY130 pKa = 9.75SPMEE134 pKa = 3.78RR135 pKa = 11.84ALIIDD140 pKa = 4.04AMASRR145 pKa = 11.84FKK147 pKa = 11.26EE148 pKa = 4.13PMFLLADD155 pKa = 3.41HH156 pKa = 6.87HH157 pKa = 7.03RR158 pKa = 11.84FDD160 pKa = 3.65AHH162 pKa = 7.03VNGPLLDD169 pKa = 3.97EE170 pKa = 3.98EE171 pKa = 4.42HH172 pKa = 7.09RR173 pKa = 11.84FYY175 pKa = 11.27LRR177 pKa = 11.84CRR179 pKa = 11.84RR180 pKa = 11.84WNLEE184 pKa = 3.61LRR186 pKa = 11.84QLLEE190 pKa = 3.86WQKK193 pKa = 10.56KK194 pKa = 8.06NKK196 pKa = 9.39GVSHH200 pKa = 6.53GGGRR204 pKa = 11.84YY205 pKa = 8.97RR206 pKa = 11.84MRR208 pKa = 11.84AKK210 pKa = 10.56RR211 pKa = 11.84MSGDD215 pKa = 3.71LNTSLGNSVINYY227 pKa = 8.31GVLKK231 pKa = 10.53AFCKK235 pKa = 10.34HH236 pKa = 5.46FNIDD240 pKa = 2.58ASMFIDD246 pKa = 4.43GDD248 pKa = 3.85DD249 pKa = 4.46SIMIMEE255 pKa = 4.69KK256 pKa = 9.82QHH258 pKa = 7.18LPDD261 pKa = 3.9LVAFAEE267 pKa = 4.27KK268 pKa = 10.19FGLVTEE274 pKa = 4.47VEE276 pKa = 4.58VVHH279 pKa = 7.08DD280 pKa = 3.48IRR282 pKa = 11.84HH283 pKa = 6.14AEE285 pKa = 4.13FCQSRR290 pKa = 11.84IIYY293 pKa = 9.51LEE295 pKa = 4.08KK296 pKa = 10.58GPIMVRR302 pKa = 11.84NPWKK306 pKa = 10.78IMDD309 pKa = 4.67CMCKK313 pKa = 9.9SPRR316 pKa = 11.84KK317 pKa = 9.66LRR319 pKa = 11.84PEE321 pKa = 3.73QARR324 pKa = 11.84GVLAATALCEE334 pKa = 4.31LMQSPGVPVIAPCASALLTYY354 pKa = 10.82AGGKK358 pKa = 9.54PMFITPTAWGKK369 pKa = 8.85FQGWQTDD376 pKa = 3.73QIVDD380 pKa = 3.52IVDD383 pKa = 3.53EE384 pKa = 4.27SARR387 pKa = 11.84ADD389 pKa = 4.46LEE391 pKa = 4.49FAWGMTISEE400 pKa = 4.12QLMMEE405 pKa = 4.51EE406 pKa = 4.88HH407 pKa = 6.19YY408 pKa = 10.91KK409 pKa = 10.4HH410 pKa = 6.02YY411 pKa = 10.73ARR413 pKa = 11.84EE414 pKa = 4.12GVTIQMPNPKK424 pKa = 9.65SKK426 pKa = 10.36PKK428 pKa = 10.22EE429 pKa = 3.7IEE431 pKa = 3.74FEE433 pKa = 3.64IWDD436 pKa = 4.22AYY438 pKa = 7.39QTQYY442 pKa = 11.25QPTEE446 pKa = 3.79VRR448 pKa = 11.84RR449 pKa = 11.84WWRR452 pKa = 11.84DD453 pKa = 2.53RR454 pKa = 11.84WEE456 pKa = 3.96ISQYY460 pKa = 10.94LPPLDD465 pKa = 4.48PEE467 pKa = 4.01AVYY470 pKa = 10.66YY471 pKa = 11.08AEE473 pKa = 4.98LLGG476 pKa = 4.35
MM1 pKa = 7.31KK2 pKa = 10.21TPDD5 pKa = 3.67PQVNSAEE12 pKa = 3.98WKK14 pKa = 9.38QFRR17 pKa = 11.84KK18 pKa = 9.9VLRR21 pKa = 11.84EE22 pKa = 3.77LSDD25 pKa = 3.59EE26 pKa = 4.28VGSVPKK32 pKa = 9.55ATPTTVVNHH41 pKa = 6.13RR42 pKa = 11.84NSMKK46 pKa = 10.06KK47 pKa = 9.61RR48 pKa = 11.84RR49 pKa = 11.84FGKK52 pKa = 10.44GMEE55 pKa = 3.76IYY57 pKa = 10.52LRR59 pKa = 11.84EE60 pKa = 4.01GVAAKK65 pKa = 10.22HH66 pKa = 6.27SYY68 pKa = 7.71ITMMPKK74 pKa = 10.38LEE76 pKa = 4.87FYY78 pKa = 10.72DD79 pKa = 3.99SEE81 pKa = 4.62KK82 pKa = 10.74IPLKK86 pKa = 9.87EE87 pKa = 4.25DD88 pKa = 2.72RR89 pKa = 11.84GIQYY93 pKa = 10.04RR94 pKa = 11.84SPVYY98 pKa = 9.31NAALARR104 pKa = 11.84HH105 pKa = 5.67LHH107 pKa = 5.59HH108 pKa = 8.33VEE110 pKa = 3.6EE111 pKa = 4.28QLYY114 pKa = 8.12KK115 pKa = 10.18TIKK118 pKa = 10.47NVDD121 pKa = 3.59GTPVFAKK128 pKa = 10.05GYY130 pKa = 9.75SPMEE134 pKa = 3.78RR135 pKa = 11.84ALIIDD140 pKa = 4.04AMASRR145 pKa = 11.84FKK147 pKa = 11.26EE148 pKa = 4.13PMFLLADD155 pKa = 3.41HH156 pKa = 6.87HH157 pKa = 7.03RR158 pKa = 11.84FDD160 pKa = 3.65AHH162 pKa = 7.03VNGPLLDD169 pKa = 3.97EE170 pKa = 3.98EE171 pKa = 4.42HH172 pKa = 7.09RR173 pKa = 11.84FYY175 pKa = 11.27LRR177 pKa = 11.84CRR179 pKa = 11.84RR180 pKa = 11.84WNLEE184 pKa = 3.61LRR186 pKa = 11.84QLLEE190 pKa = 3.86WQKK193 pKa = 10.56KK194 pKa = 8.06NKK196 pKa = 9.39GVSHH200 pKa = 6.53GGGRR204 pKa = 11.84YY205 pKa = 8.97RR206 pKa = 11.84MRR208 pKa = 11.84AKK210 pKa = 10.56RR211 pKa = 11.84MSGDD215 pKa = 3.71LNTSLGNSVINYY227 pKa = 8.31GVLKK231 pKa = 10.53AFCKK235 pKa = 10.34HH236 pKa = 5.46FNIDD240 pKa = 2.58ASMFIDD246 pKa = 4.43GDD248 pKa = 3.85DD249 pKa = 4.46SIMIMEE255 pKa = 4.69KK256 pKa = 9.82QHH258 pKa = 7.18LPDD261 pKa = 3.9LVAFAEE267 pKa = 4.27KK268 pKa = 10.19FGLVTEE274 pKa = 4.47VEE276 pKa = 4.58VVHH279 pKa = 7.08DD280 pKa = 3.48IRR282 pKa = 11.84HH283 pKa = 6.14AEE285 pKa = 4.13FCQSRR290 pKa = 11.84IIYY293 pKa = 9.51LEE295 pKa = 4.08KK296 pKa = 10.58GPIMVRR302 pKa = 11.84NPWKK306 pKa = 10.78IMDD309 pKa = 4.67CMCKK313 pKa = 9.9SPRR316 pKa = 11.84KK317 pKa = 9.66LRR319 pKa = 11.84PEE321 pKa = 3.73QARR324 pKa = 11.84GVLAATALCEE334 pKa = 4.31LMQSPGVPVIAPCASALLTYY354 pKa = 10.82AGGKK358 pKa = 9.54PMFITPTAWGKK369 pKa = 8.85FQGWQTDD376 pKa = 3.73QIVDD380 pKa = 3.52IVDD383 pKa = 3.53EE384 pKa = 4.27SARR387 pKa = 11.84ADD389 pKa = 4.46LEE391 pKa = 4.49FAWGMTISEE400 pKa = 4.12QLMMEE405 pKa = 4.51EE406 pKa = 4.88HH407 pKa = 6.19YY408 pKa = 10.91KK409 pKa = 10.4HH410 pKa = 6.02YY411 pKa = 10.73ARR413 pKa = 11.84EE414 pKa = 4.12GVTIQMPNPKK424 pKa = 9.65SKK426 pKa = 10.36PKK428 pKa = 10.22EE429 pKa = 3.7IEE431 pKa = 3.74FEE433 pKa = 3.64IWDD436 pKa = 4.22AYY438 pKa = 7.39QTQYY442 pKa = 11.25QPTEE446 pKa = 3.79VRR448 pKa = 11.84RR449 pKa = 11.84WWRR452 pKa = 11.84DD453 pKa = 2.53RR454 pKa = 11.84WEE456 pKa = 3.96ISQYY460 pKa = 10.94LPPLDD465 pKa = 4.48PEE467 pKa = 4.01AVYY470 pKa = 10.66YY471 pKa = 11.08AEE473 pKa = 4.98LLGG476 pKa = 4.35
Molecular weight: 55.15 kDa
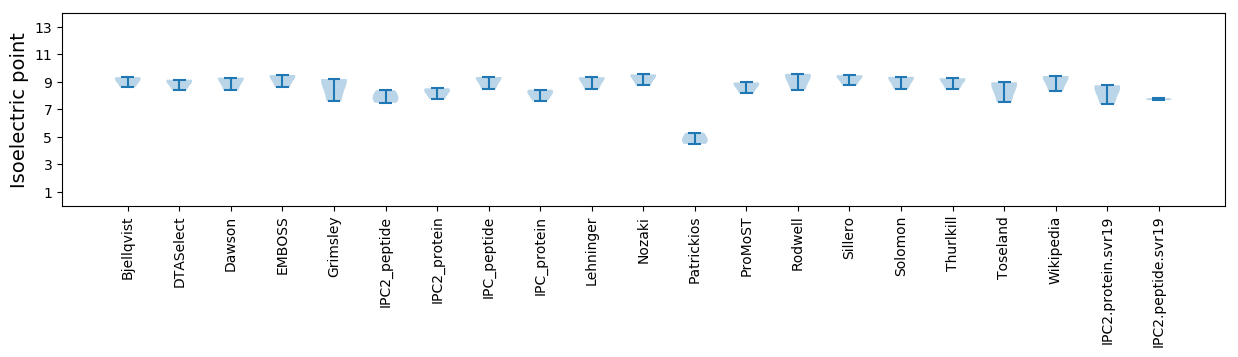
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG86|A0A1L3KG86_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 14 OX=1923260 PE=4 SV=1
MM1 pKa = 6.94FQLFEE6 pKa = 5.3SEE8 pKa = 4.31LQTCRR13 pKa = 11.84GCGNIARR20 pKa = 11.84VYY22 pKa = 10.44CEE24 pKa = 4.15KK25 pKa = 10.58CRR27 pKa = 11.84QSYY30 pKa = 10.76AKK32 pKa = 10.38YY33 pKa = 10.53DD34 pKa = 3.93LNTKK38 pKa = 9.22EE39 pKa = 3.86RR40 pKa = 11.84LGRR43 pKa = 11.84RR44 pKa = 11.84SYY46 pKa = 8.8WCRR49 pKa = 11.84AKK51 pKa = 10.4RR52 pKa = 11.84SHH54 pKa = 6.97PLAEE58 pKa = 3.87EE59 pKa = 4.44AIYY62 pKa = 10.45RR63 pKa = 11.84PEE65 pKa = 4.35AQIPWCEE72 pKa = 4.36CKK74 pKa = 10.54RR75 pKa = 11.84LDD77 pKa = 4.07GSPMPGLQEE86 pKa = 4.17KK87 pKa = 9.36QQQQQQQAQQQPQQQTYY104 pKa = 10.92APIQPISEE112 pKa = 4.2QKK114 pKa = 10.32LAAAKK119 pKa = 9.85AQAAASTTTSQQQQIRR135 pKa = 11.84DD136 pKa = 3.67EE137 pKa = 4.61HH138 pKa = 6.0GTWPTKK144 pKa = 9.91SAARR148 pKa = 11.84QAHH151 pKa = 4.64YY152 pKa = 9.5QARR155 pKa = 11.84AVGWRR160 pKa = 11.84EE161 pKa = 3.64LQRR164 pKa = 11.84EE165 pKa = 4.21KK166 pKa = 11.26NKK168 pKa = 10.12ARR170 pKa = 11.84RR171 pKa = 11.84QEE173 pKa = 4.17SKK175 pKa = 10.82VLSGTQSMGKK185 pKa = 8.98QRR187 pKa = 11.84PFRR190 pKa = 11.84APPAVSPVRR199 pKa = 11.84GQRR202 pKa = 11.84FEE204 pKa = 4.08QVQCASCGGSGSMWLYY220 pKa = 10.25QPIYY224 pKa = 10.75RR225 pKa = 11.84NDD227 pKa = 3.37PPYY230 pKa = 11.3NRR232 pKa = 11.84GGAKK236 pKa = 10.32AKK238 pKa = 9.91GQRR241 pKa = 11.84LDD243 pKa = 3.25QTAAASTTNQQQQHH257 pKa = 5.02QQQQQQPSSPYY268 pKa = 7.9TKK270 pKa = 10.31AFLRR274 pKa = 11.84RR275 pKa = 11.84EE276 pKa = 3.46RR277 pKa = 11.84RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84QKK282 pKa = 10.4QYY284 pKa = 9.91YY285 pKa = 8.85EE286 pKa = 3.85EE287 pKa = 4.25EE288 pKa = 3.65ARR290 pKa = 11.84QRR292 pKa = 11.84LEE294 pKa = 4.4GMGQRR299 pKa = 11.84LGNPEE304 pKa = 3.88QNDD307 pKa = 3.48EE308 pKa = 4.28RR309 pKa = 11.84DD310 pKa = 3.54QPPAGQSEE318 pKa = 4.43EE319 pKa = 3.77QGGIRR324 pKa = 11.84EE325 pKa = 4.35CRR327 pKa = 11.84VEE329 pKa = 4.49EE330 pKa = 4.25ATCPDD335 pKa = 3.85AGASDD340 pKa = 3.76IAPRR344 pKa = 11.84CPSKK348 pKa = 11.13SPARR352 pKa = 11.84CGGSPGAKK360 pKa = 9.22PSTDD364 pKa = 2.86RR365 pKa = 11.84TSEE368 pKa = 4.16SNPRR372 pKa = 11.84SASPEE377 pKa = 3.78ARR379 pKa = 11.84PAPCSALAGEE389 pKa = 4.65HH390 pKa = 6.54GEE392 pKa = 4.07PLALPGASEE401 pKa = 4.17QPKK404 pKa = 10.14SSRR407 pKa = 11.84GNQRR411 pKa = 11.84TRR413 pKa = 11.84RR414 pKa = 11.84NRR416 pKa = 11.84RR417 pKa = 11.84KK418 pKa = 9.35IAATLPHH425 pKa = 6.32QPHH428 pKa = 6.54SPTKK432 pKa = 9.62GSAGKK437 pKa = 10.12DD438 pKa = 3.06VLAPSDD444 pKa = 3.8EE445 pKa = 4.66GCGDD449 pKa = 3.41RR450 pKa = 11.84YY451 pKa = 11.15DD452 pKa = 3.53RR453 pKa = 11.84AIRR456 pKa = 11.84QLGSVEE462 pKa = 4.13QPGSEE467 pKa = 4.22SEE469 pKa = 4.29GGFDD473 pKa = 4.91EE474 pKa = 5.33PEE476 pKa = 3.81VDD478 pKa = 5.4AKK480 pKa = 11.34SCDD483 pKa = 3.39GRR485 pKa = 11.84DD486 pKa = 3.14
MM1 pKa = 6.94FQLFEE6 pKa = 5.3SEE8 pKa = 4.31LQTCRR13 pKa = 11.84GCGNIARR20 pKa = 11.84VYY22 pKa = 10.44CEE24 pKa = 4.15KK25 pKa = 10.58CRR27 pKa = 11.84QSYY30 pKa = 10.76AKK32 pKa = 10.38YY33 pKa = 10.53DD34 pKa = 3.93LNTKK38 pKa = 9.22EE39 pKa = 3.86RR40 pKa = 11.84LGRR43 pKa = 11.84RR44 pKa = 11.84SYY46 pKa = 8.8WCRR49 pKa = 11.84AKK51 pKa = 10.4RR52 pKa = 11.84SHH54 pKa = 6.97PLAEE58 pKa = 3.87EE59 pKa = 4.44AIYY62 pKa = 10.45RR63 pKa = 11.84PEE65 pKa = 4.35AQIPWCEE72 pKa = 4.36CKK74 pKa = 10.54RR75 pKa = 11.84LDD77 pKa = 4.07GSPMPGLQEE86 pKa = 4.17KK87 pKa = 9.36QQQQQQQAQQQPQQQTYY104 pKa = 10.92APIQPISEE112 pKa = 4.2QKK114 pKa = 10.32LAAAKK119 pKa = 9.85AQAAASTTTSQQQQIRR135 pKa = 11.84DD136 pKa = 3.67EE137 pKa = 4.61HH138 pKa = 6.0GTWPTKK144 pKa = 9.91SAARR148 pKa = 11.84QAHH151 pKa = 4.64YY152 pKa = 9.5QARR155 pKa = 11.84AVGWRR160 pKa = 11.84EE161 pKa = 3.64LQRR164 pKa = 11.84EE165 pKa = 4.21KK166 pKa = 11.26NKK168 pKa = 10.12ARR170 pKa = 11.84RR171 pKa = 11.84QEE173 pKa = 4.17SKK175 pKa = 10.82VLSGTQSMGKK185 pKa = 8.98QRR187 pKa = 11.84PFRR190 pKa = 11.84APPAVSPVRR199 pKa = 11.84GQRR202 pKa = 11.84FEE204 pKa = 4.08QVQCASCGGSGSMWLYY220 pKa = 10.25QPIYY224 pKa = 10.75RR225 pKa = 11.84NDD227 pKa = 3.37PPYY230 pKa = 11.3NRR232 pKa = 11.84GGAKK236 pKa = 10.32AKK238 pKa = 9.91GQRR241 pKa = 11.84LDD243 pKa = 3.25QTAAASTTNQQQQHH257 pKa = 5.02QQQQQQPSSPYY268 pKa = 7.9TKK270 pKa = 10.31AFLRR274 pKa = 11.84RR275 pKa = 11.84EE276 pKa = 3.46RR277 pKa = 11.84RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84QKK282 pKa = 10.4QYY284 pKa = 9.91YY285 pKa = 8.85EE286 pKa = 3.85EE287 pKa = 4.25EE288 pKa = 3.65ARR290 pKa = 11.84QRR292 pKa = 11.84LEE294 pKa = 4.4GMGQRR299 pKa = 11.84LGNPEE304 pKa = 3.88QNDD307 pKa = 3.48EE308 pKa = 4.28RR309 pKa = 11.84DD310 pKa = 3.54QPPAGQSEE318 pKa = 4.43EE319 pKa = 3.77QGGIRR324 pKa = 11.84EE325 pKa = 4.35CRR327 pKa = 11.84VEE329 pKa = 4.49EE330 pKa = 4.25ATCPDD335 pKa = 3.85AGASDD340 pKa = 3.76IAPRR344 pKa = 11.84CPSKK348 pKa = 11.13SPARR352 pKa = 11.84CGGSPGAKK360 pKa = 9.22PSTDD364 pKa = 2.86RR365 pKa = 11.84TSEE368 pKa = 4.16SNPRR372 pKa = 11.84SASPEE377 pKa = 3.78ARR379 pKa = 11.84PAPCSALAGEE389 pKa = 4.65HH390 pKa = 6.54GEE392 pKa = 4.07PLALPGASEE401 pKa = 4.17QPKK404 pKa = 10.14SSRR407 pKa = 11.84GNQRR411 pKa = 11.84TRR413 pKa = 11.84RR414 pKa = 11.84NRR416 pKa = 11.84RR417 pKa = 11.84KK418 pKa = 9.35IAATLPHH425 pKa = 6.32QPHH428 pKa = 6.54SPTKK432 pKa = 9.62GSAGKK437 pKa = 10.12DD438 pKa = 3.06VLAPSDD444 pKa = 3.8EE445 pKa = 4.66GCGDD449 pKa = 3.41RR450 pKa = 11.84YY451 pKa = 11.15DD452 pKa = 3.53RR453 pKa = 11.84AIRR456 pKa = 11.84QLGSVEE462 pKa = 4.13QPGSEE467 pKa = 4.22SEE469 pKa = 4.29GGFDD473 pKa = 4.91EE474 pKa = 5.33PEE476 pKa = 3.81VDD478 pKa = 5.4AKK480 pKa = 11.34SCDD483 pKa = 3.39GRR485 pKa = 11.84DD486 pKa = 3.14
Molecular weight: 54.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1649 |
476 |
687 |
549.7 |
62.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.126 ± 0.831 | 2.183 ± 0.387 |
4.366 ± 0.27 | 7.641 ± 0.112 |
2.365 ± 0.53 | 6.125 ± 0.685 |
2.608 ± 0.407 | 3.457 ± 0.724 |
6.367 ± 0.495 | 6.186 ± 0.698 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.79 ± 0.726 | 4.73 ± 1.534 |
6.853 ± 0.488 | 8.187 ± 1.685 |
8.126 ± 0.813 | 6.61 ± 0.691 |
4.669 ± 0.597 | 4.548 ± 0.864 |
1.455 ± 0.309 | 2.608 ± 0.65 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
