
Lysobacter daejeonensis GH1-9
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Lysobacter; Lysobacter daejeonensis
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
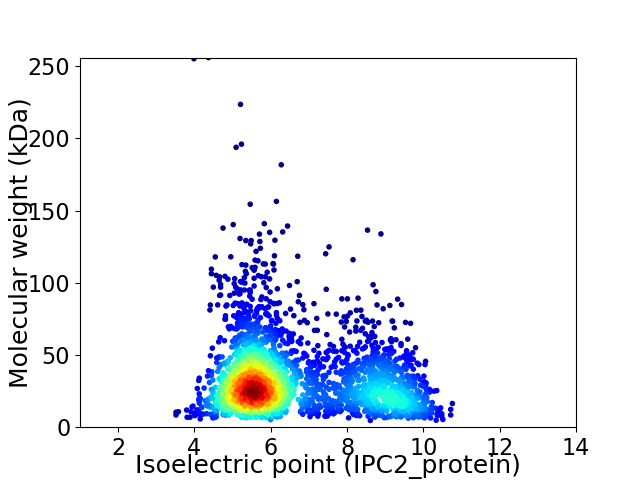
Virtual 2D-PAGE plot for 2570 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
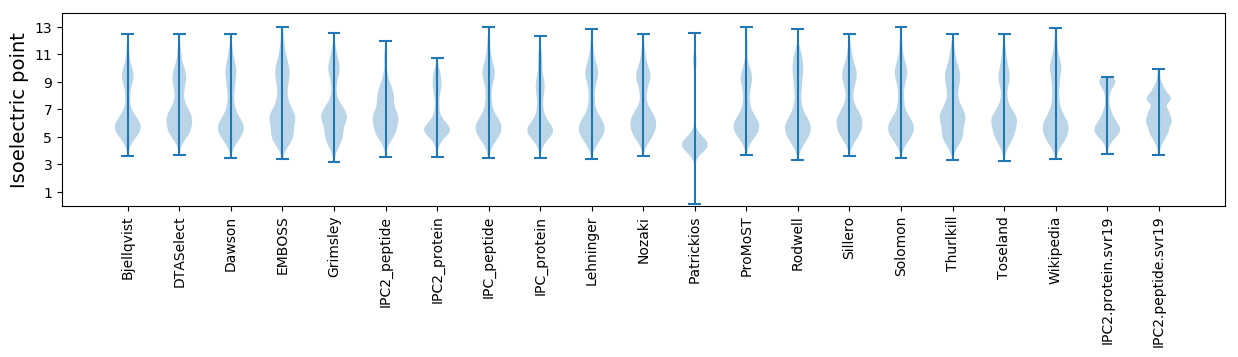
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0ETD8|A0A0A0ETD8_9GAMM LacI family transcriptional regulator OS=Lysobacter daejeonensis GH1-9 OX=1385517 GN=N800_07390 PE=4 SV=1
MM1 pKa = 7.72ALACLLLMVSATPAAQAQTVTATNSLSCLANRR33 pKa = 11.84MDD35 pKa = 3.67NPTNLGCTANDD46 pKa = 3.68FSHH49 pKa = 7.05IVTAQALSAPTTCTAGSEE67 pKa = 4.22ITVSVQLTLQSNSTDD82 pKa = 3.34RR83 pKa = 11.84YY84 pKa = 10.89DD85 pKa = 3.18IGLFMGEE92 pKa = 4.89DD93 pKa = 3.95GNNPAQLGGTCSVATFPEE111 pKa = 4.81SPAPFADD118 pKa = 4.33LDD120 pKa = 4.25AGTQPTDD127 pKa = 2.79TCGDD131 pKa = 3.83FNANGSATVTVQNVKK146 pKa = 8.69LTCTAGADD154 pKa = 3.89GKK156 pKa = 11.16LVMPYY161 pKa = 9.69TLAYY165 pKa = 9.03SQNTGGVCTGPGVVRR180 pKa = 11.84AGTTSKK186 pKa = 9.87CTGGTASVSNIVVAGHH202 pKa = 6.26VDD204 pKa = 3.45VTKK207 pKa = 9.62QTLPDD212 pKa = 3.85AAPDD216 pKa = 3.7SFAFTAVDD224 pKa = 4.66GPDD227 pKa = 3.34TSPTSFNLVDD237 pKa = 3.94GGTQRR242 pKa = 11.84VTTTFGATGSSITIDD257 pKa = 3.64EE258 pKa = 4.43NVPPHH263 pKa = 6.12WRR265 pKa = 11.84SSVACASSNVDD276 pKa = 2.98TSGQANGTFIANLTSAEE293 pKa = 4.02PTANCTVTNTKK304 pKa = 10.09RR305 pKa = 11.84SRR307 pKa = 11.84ITLVKK312 pKa = 10.57NIVGGRR318 pKa = 11.84NNAADD323 pKa = 3.63QFTVSGTGGGTLTNTSDD340 pKa = 3.58VTMSAPVSATTSGTSTTASVTFYY363 pKa = 10.82SSPGSGLTLTDD374 pKa = 3.04AMAAGSVSTLADD386 pKa = 3.34YY387 pKa = 9.7TPTYY391 pKa = 10.68SCTNATAGSPTSMPSGSGATFGLTPAPGDD420 pKa = 5.15DD421 pKa = 3.52ITCTYY426 pKa = 10.3TNTLIARR433 pKa = 11.84PSLLLDD439 pKa = 3.32KK440 pKa = 10.66RR441 pKa = 11.84LVSGAGFDD449 pKa = 3.74SVGDD453 pKa = 4.05VIQYY457 pKa = 10.88AYY459 pKa = 10.84DD460 pKa = 3.47VTNSGNVTITGPITVSDD477 pKa = 4.13DD478 pKa = 3.62KK479 pKa = 10.9IASVTCPAGDD489 pKa = 4.08LAPGAMITCTASYY502 pKa = 9.49TVAQADD508 pKa = 3.81INNGSVTNVATATGSFNNQPITSNADD534 pKa = 3.36TVTVEE539 pKa = 4.15ALQEE543 pKa = 3.98PALTLAKK550 pKa = 9.32TAVSGDD556 pKa = 3.88PYY558 pKa = 11.41DD559 pKa = 4.46SIGDD563 pKa = 3.81VVQYY567 pKa = 10.51EE568 pKa = 4.29YY569 pKa = 11.44LVTNSGNVVITDD581 pKa = 5.42PITVADD587 pKa = 4.67DD588 pKa = 3.66KK589 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY614 pKa = 9.82TITQADD620 pKa = 3.37VDD622 pKa = 4.25AGSVTNLATATSGTVNSNQATEE644 pKa = 4.4TVNATRR650 pKa = 11.84SPSLGLEE657 pKa = 4.18KK658 pKa = 9.88TASPLVYY665 pKa = 10.05AAAGDD670 pKa = 4.15VISYY674 pKa = 10.61SYY676 pKa = 11.63LLTNTGNVTLSAPYY690 pKa = 9.2TVADD694 pKa = 4.41DD695 pKa = 4.2KK696 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY720 pKa = 9.36TITQTDD726 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD752 pKa = 3.58SATVTATQTPTLSLDD767 pKa = 3.06KK768 pKa = 10.63RR769 pKa = 11.84LVSGAGFDD777 pKa = 3.74SVGDD781 pKa = 4.05VIQYY785 pKa = 10.88AYY787 pKa = 10.84DD788 pKa = 3.47VTNSGNVTITGPITVSDD805 pKa = 4.13DD806 pKa = 3.62KK807 pKa = 10.9IASVTCPAGDD817 pKa = 4.08LAPGAMITCTASYY830 pKa = 9.49TVAQADD836 pKa = 3.81INNGSVTNVATATGSFNNQPITSNADD862 pKa = 3.36TVTVEE867 pKa = 4.15ALQEE871 pKa = 3.98PALTLAKK878 pKa = 9.32TAVSGDD884 pKa = 3.88PYY886 pKa = 11.41DD887 pKa = 4.46SIGDD891 pKa = 3.81VVQYY895 pKa = 10.51EE896 pKa = 4.29YY897 pKa = 11.44LVTNSGNVVITDD909 pKa = 5.42PITVADD915 pKa = 4.67DD916 pKa = 3.66KK917 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY942 pKa = 9.82TITQADD948 pKa = 3.37VDD950 pKa = 4.25AGSVTNLATATSGTVNSNQATEE972 pKa = 4.4TVNATRR978 pKa = 11.84SPSLGLEE985 pKa = 4.18KK986 pKa = 9.88TASPLVYY993 pKa = 10.05AAAGDD998 pKa = 4.15VISYY1002 pKa = 10.61SYY1004 pKa = 11.63LLTNTGNVTLSAPYY1018 pKa = 9.2TVADD1022 pKa = 4.41DD1023 pKa = 4.2KK1024 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1048 pKa = 9.36TITQTDD1054 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1080 pKa = 3.58SATVTATQTPTLSLDD1095 pKa = 3.06KK1096 pKa = 10.63RR1097 pKa = 11.84LVSGAGFDD1105 pKa = 3.74SVGDD1109 pKa = 4.05VIQYY1113 pKa = 10.88AYY1115 pKa = 10.84DD1116 pKa = 3.47VTNSGNVTITGPITISDD1133 pKa = 3.9DD1134 pKa = 3.72KK1135 pKa = 10.6IASVTCPAGDD1145 pKa = 4.08LAPGAMVTCTASYY1158 pKa = 9.77TVTQADD1164 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1190 pKa = 3.38TVTVEE1195 pKa = 4.06ALQEE1199 pKa = 3.98PALTLAKK1206 pKa = 9.32TAVSGDD1212 pKa = 3.95PYY1214 pKa = 11.28DD1215 pKa = 3.51SVGDD1219 pKa = 3.88VVQYY1223 pKa = 10.7EE1224 pKa = 4.27YY1225 pKa = 11.44LVTNSGNVVITDD1237 pKa = 5.42PITVADD1243 pKa = 4.67DD1244 pKa = 3.66KK1245 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY1270 pKa = 9.82TITQADD1276 pKa = 3.37VDD1278 pKa = 4.25AGSVTNLATATSGTVNSNQATEE1300 pKa = 4.4TVNATRR1306 pKa = 11.84SPSLGLEE1313 pKa = 4.18KK1314 pKa = 9.88TASPLVYY1321 pKa = 10.05AAAGDD1326 pKa = 4.15VISYY1330 pKa = 10.61SYY1332 pKa = 11.63LLTNTGNVTLSAPYY1346 pKa = 9.2TVADD1350 pKa = 4.41DD1351 pKa = 4.2KK1352 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1376 pKa = 9.36TITQTDD1382 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1408 pKa = 3.58SATVTATQTPTLSLDD1423 pKa = 3.06KK1424 pKa = 10.63RR1425 pKa = 11.84LVSGAGFDD1433 pKa = 3.74SVGDD1437 pKa = 4.05VIQYY1441 pKa = 10.88AYY1443 pKa = 10.84DD1444 pKa = 3.47VTNSGNVTITGPITISDD1461 pKa = 3.9DD1462 pKa = 3.72KK1463 pKa = 10.6IASVTCPAGDD1473 pKa = 4.08LAPGAMVTCTASYY1486 pKa = 9.77TVTQADD1492 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1518 pKa = 3.38TVTVEE1523 pKa = 4.06ALQEE1527 pKa = 3.87PALTLDD1533 pKa = 3.41KK1534 pKa = 10.69RR1535 pKa = 11.84VVSGNPYY1542 pKa = 10.64DD1543 pKa = 3.65SVGDD1547 pKa = 3.93VVQYY1551 pKa = 10.6TYY1553 pKa = 11.62DD1554 pKa = 3.42VTNTGNLTITDD1565 pKa = 5.9PITVSDD1571 pKa = 4.4DD1572 pKa = 3.58KK1573 pKa = 11.08IATVNCPALPSGGLAPGAMLTCTASYY1599 pKa = 9.8TITQADD1605 pKa = 3.37VDD1607 pKa = 4.25AGSVTNLATATSGTVNSNQATEE1629 pKa = 4.4TVNATRR1635 pKa = 11.84SPSLGLEE1642 pKa = 4.18KK1643 pKa = 9.88TASPLVYY1650 pKa = 10.05AAAGDD1655 pKa = 4.15VISYY1659 pKa = 10.61SYY1661 pKa = 11.63LLTNTGNVTLSAPYY1675 pKa = 9.2TVADD1679 pKa = 4.41DD1680 pKa = 4.2KK1681 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1705 pKa = 9.36TITQTDD1711 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1737 pKa = 3.58SATVTATQTPTLSLDD1752 pKa = 3.06KK1753 pKa = 10.63RR1754 pKa = 11.84LVSGAGFDD1762 pKa = 3.74SVGDD1766 pKa = 4.05VIQYY1770 pKa = 10.88AYY1772 pKa = 10.84DD1773 pKa = 3.47VTNSGNVTITGPITISDD1790 pKa = 3.9DD1791 pKa = 3.72KK1792 pKa = 10.6IASVTCPAGDD1802 pKa = 4.08LAPGAMVTCTASYY1815 pKa = 9.77TVTQADD1821 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1847 pKa = 3.38TVTVEE1852 pKa = 4.06ALQEE1856 pKa = 3.87PALTLDD1862 pKa = 3.41KK1863 pKa = 10.69RR1864 pKa = 11.84VVSGNPYY1871 pKa = 10.64DD1872 pKa = 3.65SVGDD1876 pKa = 3.93VVQYY1880 pKa = 10.6TYY1882 pKa = 11.62DD1883 pKa = 3.42VTNTGNLTITDD1894 pKa = 5.9PITVSDD1900 pKa = 4.4DD1901 pKa = 3.58KK1902 pKa = 11.08IATVNCPALPSGGLAPGAMLTCTASYY1928 pKa = 9.84TITQADD1934 pKa = 3.82INNGSVTNVATASSGTTQSPPDD1956 pKa = 4.09TVTVTATQRR1965 pKa = 11.84PALSLDD1971 pKa = 2.97KK1972 pKa = 10.66RR1973 pKa = 11.84LLSGTTVTAVGDD1985 pKa = 4.18VIQYY1989 pKa = 10.88AYY1991 pKa = 10.66DD1992 pKa = 3.38VRR1994 pKa = 11.84NTGNVTITGPITVSDD2009 pKa = 4.08DD2010 pKa = 3.64KK2011 pKa = 11.05IATVTCPAGDD2021 pKa = 4.1LAPAAMLTCTASYY2034 pKa = 9.83TVTQADD2040 pKa = 4.31LDD2042 pKa = 4.2AGSITNIATASGTFNEE2058 pKa = 4.34QTVTSNQDD2066 pKa = 3.09SVTVEE2071 pKa = 4.36ALHH2074 pKa = 6.21EE2075 pKa = 4.14MAVVLTKK2082 pKa = 10.23IAAKK2086 pKa = 10.32RR2087 pKa = 11.84EE2088 pKa = 3.99VRR2090 pKa = 11.84VGDD2093 pKa = 3.5LVRR2096 pKa = 11.84FTVVARR2102 pKa = 11.84NVGPGPVVDD2111 pKa = 4.23ATLIDD2116 pKa = 3.69TPPRR2120 pKa = 11.84GFTFVEE2126 pKa = 5.03GSLTADD2132 pKa = 3.93DD2133 pKa = 4.69EE2134 pKa = 4.99DD2135 pKa = 5.89DD2136 pKa = 3.81NVVVSGVDD2144 pKa = 3.7PIRR2147 pKa = 11.84ISGVDD2152 pKa = 2.82IATNGTATFVYY2163 pKa = 9.6FLRR2166 pKa = 11.84VGAGVGHH2173 pKa = 6.22GTHH2176 pKa = 6.85INRR2179 pKa = 11.84VIAVDD2184 pKa = 3.75PTEE2187 pKa = 4.06HH2188 pKa = 5.81TVSNEE2193 pKa = 3.11AAAEE2197 pKa = 4.1VVIIGDD2203 pKa = 3.69PMLDD2207 pKa = 3.1EE2208 pKa = 4.38SLIVGTVFDD2217 pKa = 5.08DD2218 pKa = 4.0RR2219 pKa = 11.84NGDD2222 pKa = 4.27GIQQPGEE2229 pKa = 3.78RR2230 pKa = 11.84GIPGVRR2236 pKa = 11.84IGSVEE2241 pKa = 3.96GLIMEE2246 pKa = 4.67TDD2248 pKa = 2.98AFGRR2252 pKa = 11.84YY2253 pKa = 8.87HH2254 pKa = 7.17LVGIDD2259 pKa = 3.19GGDD2262 pKa = 3.82FSRR2265 pKa = 11.84GRR2267 pKa = 11.84NFILKK2272 pKa = 9.58VDD2274 pKa = 3.96ASTLPVGTEE2283 pKa = 3.9FTTEE2287 pKa = 3.55NPRR2290 pKa = 11.84VRR2292 pKa = 11.84RR2293 pKa = 11.84ITPGVPVRR2301 pKa = 11.84FDD2303 pKa = 3.65FGVKK2307 pKa = 10.25LPNGEE2312 pKa = 4.21MKK2314 pKa = 10.61GGRR2317 pKa = 11.84QEE2319 pKa = 4.53LDD2321 pKa = 3.2AEE2323 pKa = 4.48LGEE2326 pKa = 4.58VMFDD2330 pKa = 3.63PRR2332 pKa = 11.84SSTLKK2337 pKa = 10.78AEE2339 pKa = 4.1YY2340 pKa = 10.44LPVIDD2345 pKa = 5.05KK2346 pKa = 10.39VASKK2350 pKa = 10.37VRR2352 pKa = 11.84EE2353 pKa = 4.09YY2354 pKa = 11.14NGGRR2358 pKa = 11.84IVITAKK2364 pKa = 10.81AEE2366 pKa = 4.06EE2367 pKa = 4.19EE2368 pKa = 3.88ALAYY2372 pKa = 10.31ARR2374 pKa = 11.84ARR2376 pKa = 11.84AVQDD2380 pKa = 3.36ALRR2383 pKa = 11.84TKK2385 pKa = 10.76LEE2387 pKa = 3.91PALADD2392 pKa = 3.56RR2393 pKa = 11.84VAVEE2397 pKa = 4.45VVADD2401 pKa = 3.71TSGTMVRR2408 pKa = 11.84LDD2410 pKa = 3.55RR2411 pKa = 11.84GIHH2414 pKa = 6.1LGTFLFDD2421 pKa = 3.48TDD2423 pKa = 3.56KK2424 pKa = 11.54SVIKK2428 pKa = 9.46PQYY2431 pKa = 9.14RR2432 pKa = 11.84GLIKK2436 pKa = 10.57EE2437 pKa = 4.18IADD2440 pKa = 3.52DD2441 pKa = 4.31LNRR2444 pKa = 11.84MGRR2447 pKa = 11.84GTIGLVGHH2455 pKa = 7.23ADD2457 pKa = 3.06MRR2459 pKa = 11.84ASNEE2463 pKa = 3.67YY2464 pKa = 10.28NRR2466 pKa = 11.84YY2467 pKa = 9.65LGLRR2471 pKa = 11.84RR2472 pKa = 11.84SKK2474 pKa = 11.19AVFDD2478 pKa = 5.45AILAQLRR2485 pKa = 11.84PEE2487 pKa = 3.76VRR2489 pKa = 11.84QRR2491 pKa = 11.84VRR2493 pKa = 11.84VDD2495 pKa = 2.73ISDD2498 pKa = 4.45EE2499 pKa = 3.86IDD2501 pKa = 3.36AEE2503 pKa = 4.42TGVGDD2508 pKa = 3.73RR2509 pKa = 4.83
MM1 pKa = 7.72ALACLLLMVSATPAAQAQTVTATNSLSCLANRR33 pKa = 11.84MDD35 pKa = 3.67NPTNLGCTANDD46 pKa = 3.68FSHH49 pKa = 7.05IVTAQALSAPTTCTAGSEE67 pKa = 4.22ITVSVQLTLQSNSTDD82 pKa = 3.34RR83 pKa = 11.84YY84 pKa = 10.89DD85 pKa = 3.18IGLFMGEE92 pKa = 4.89DD93 pKa = 3.95GNNPAQLGGTCSVATFPEE111 pKa = 4.81SPAPFADD118 pKa = 4.33LDD120 pKa = 4.25AGTQPTDD127 pKa = 2.79TCGDD131 pKa = 3.83FNANGSATVTVQNVKK146 pKa = 8.69LTCTAGADD154 pKa = 3.89GKK156 pKa = 11.16LVMPYY161 pKa = 9.69TLAYY165 pKa = 9.03SQNTGGVCTGPGVVRR180 pKa = 11.84AGTTSKK186 pKa = 9.87CTGGTASVSNIVVAGHH202 pKa = 6.26VDD204 pKa = 3.45VTKK207 pKa = 9.62QTLPDD212 pKa = 3.85AAPDD216 pKa = 3.7SFAFTAVDD224 pKa = 4.66GPDD227 pKa = 3.34TSPTSFNLVDD237 pKa = 3.94GGTQRR242 pKa = 11.84VTTTFGATGSSITIDD257 pKa = 3.64EE258 pKa = 4.43NVPPHH263 pKa = 6.12WRR265 pKa = 11.84SSVACASSNVDD276 pKa = 2.98TSGQANGTFIANLTSAEE293 pKa = 4.02PTANCTVTNTKK304 pKa = 10.09RR305 pKa = 11.84SRR307 pKa = 11.84ITLVKK312 pKa = 10.57NIVGGRR318 pKa = 11.84NNAADD323 pKa = 3.63QFTVSGTGGGTLTNTSDD340 pKa = 3.58VTMSAPVSATTSGTSTTASVTFYY363 pKa = 10.82SSPGSGLTLTDD374 pKa = 3.04AMAAGSVSTLADD386 pKa = 3.34YY387 pKa = 9.7TPTYY391 pKa = 10.68SCTNATAGSPTSMPSGSGATFGLTPAPGDD420 pKa = 5.15DD421 pKa = 3.52ITCTYY426 pKa = 10.3TNTLIARR433 pKa = 11.84PSLLLDD439 pKa = 3.32KK440 pKa = 10.66RR441 pKa = 11.84LVSGAGFDD449 pKa = 3.74SVGDD453 pKa = 4.05VIQYY457 pKa = 10.88AYY459 pKa = 10.84DD460 pKa = 3.47VTNSGNVTITGPITVSDD477 pKa = 4.13DD478 pKa = 3.62KK479 pKa = 10.9IASVTCPAGDD489 pKa = 4.08LAPGAMITCTASYY502 pKa = 9.49TVAQADD508 pKa = 3.81INNGSVTNVATATGSFNNQPITSNADD534 pKa = 3.36TVTVEE539 pKa = 4.15ALQEE543 pKa = 3.98PALTLAKK550 pKa = 9.32TAVSGDD556 pKa = 3.88PYY558 pKa = 11.41DD559 pKa = 4.46SIGDD563 pKa = 3.81VVQYY567 pKa = 10.51EE568 pKa = 4.29YY569 pKa = 11.44LVTNSGNVVITDD581 pKa = 5.42PITVADD587 pKa = 4.67DD588 pKa = 3.66KK589 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY614 pKa = 9.82TITQADD620 pKa = 3.37VDD622 pKa = 4.25AGSVTNLATATSGTVNSNQATEE644 pKa = 4.4TVNATRR650 pKa = 11.84SPSLGLEE657 pKa = 4.18KK658 pKa = 9.88TASPLVYY665 pKa = 10.05AAAGDD670 pKa = 4.15VISYY674 pKa = 10.61SYY676 pKa = 11.63LLTNTGNVTLSAPYY690 pKa = 9.2TVADD694 pKa = 4.41DD695 pKa = 4.2KK696 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY720 pKa = 9.36TITQTDD726 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD752 pKa = 3.58SATVTATQTPTLSLDD767 pKa = 3.06KK768 pKa = 10.63RR769 pKa = 11.84LVSGAGFDD777 pKa = 3.74SVGDD781 pKa = 4.05VIQYY785 pKa = 10.88AYY787 pKa = 10.84DD788 pKa = 3.47VTNSGNVTITGPITVSDD805 pKa = 4.13DD806 pKa = 3.62KK807 pKa = 10.9IASVTCPAGDD817 pKa = 4.08LAPGAMITCTASYY830 pKa = 9.49TVAQADD836 pKa = 3.81INNGSVTNVATATGSFNNQPITSNADD862 pKa = 3.36TVTVEE867 pKa = 4.15ALQEE871 pKa = 3.98PALTLAKK878 pKa = 9.32TAVSGDD884 pKa = 3.88PYY886 pKa = 11.41DD887 pKa = 4.46SIGDD891 pKa = 3.81VVQYY895 pKa = 10.51EE896 pKa = 4.29YY897 pKa = 11.44LVTNSGNVVITDD909 pKa = 5.42PITVADD915 pKa = 4.67DD916 pKa = 3.66KK917 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY942 pKa = 9.82TITQADD948 pKa = 3.37VDD950 pKa = 4.25AGSVTNLATATSGTVNSNQATEE972 pKa = 4.4TVNATRR978 pKa = 11.84SPSLGLEE985 pKa = 4.18KK986 pKa = 9.88TASPLVYY993 pKa = 10.05AAAGDD998 pKa = 4.15VISYY1002 pKa = 10.61SYY1004 pKa = 11.63LLTNTGNVTLSAPYY1018 pKa = 9.2TVADD1022 pKa = 4.41DD1023 pKa = 4.2KK1024 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1048 pKa = 9.36TITQTDD1054 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1080 pKa = 3.58SATVTATQTPTLSLDD1095 pKa = 3.06KK1096 pKa = 10.63RR1097 pKa = 11.84LVSGAGFDD1105 pKa = 3.74SVGDD1109 pKa = 4.05VIQYY1113 pKa = 10.88AYY1115 pKa = 10.84DD1116 pKa = 3.47VTNSGNVTITGPITISDD1133 pKa = 3.9DD1134 pKa = 3.72KK1135 pKa = 10.6IASVTCPAGDD1145 pKa = 4.08LAPGAMVTCTASYY1158 pKa = 9.77TVTQADD1164 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1190 pKa = 3.38TVTVEE1195 pKa = 4.06ALQEE1199 pKa = 3.98PALTLAKK1206 pKa = 9.32TAVSGDD1212 pKa = 3.95PYY1214 pKa = 11.28DD1215 pKa = 3.51SVGDD1219 pKa = 3.88VVQYY1223 pKa = 10.7EE1224 pKa = 4.27YY1225 pKa = 11.44LVTNSGNVVITDD1237 pKa = 5.42PITVADD1243 pKa = 4.67DD1244 pKa = 3.66KK1245 pKa = 10.61TTVTCPALPGGGLAPGGSITCTASYY1270 pKa = 9.82TITQADD1276 pKa = 3.37VDD1278 pKa = 4.25AGSVTNLATATSGTVNSNQATEE1300 pKa = 4.4TVNATRR1306 pKa = 11.84SPSLGLEE1313 pKa = 4.18KK1314 pKa = 9.88TASPLVYY1321 pKa = 10.05AAAGDD1326 pKa = 4.15VISYY1330 pKa = 10.61SYY1332 pKa = 11.63LLTNTGNVTLSAPYY1346 pKa = 9.2TVADD1350 pKa = 4.41DD1351 pKa = 4.2KK1352 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1376 pKa = 9.36TITQTDD1382 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1408 pKa = 3.58SATVTATQTPTLSLDD1423 pKa = 3.06KK1424 pKa = 10.63RR1425 pKa = 11.84LVSGAGFDD1433 pKa = 3.74SVGDD1437 pKa = 4.05VIQYY1441 pKa = 10.88AYY1443 pKa = 10.84DD1444 pKa = 3.47VTNSGNVTITGPITISDD1461 pKa = 3.9DD1462 pKa = 3.72KK1463 pKa = 10.6IASVTCPAGDD1473 pKa = 4.08LAPGAMVTCTASYY1486 pKa = 9.77TVTQADD1492 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1518 pKa = 3.38TVTVEE1523 pKa = 4.06ALQEE1527 pKa = 3.87PALTLDD1533 pKa = 3.41KK1534 pKa = 10.69RR1535 pKa = 11.84VVSGNPYY1542 pKa = 10.64DD1543 pKa = 3.65SVGDD1547 pKa = 3.93VVQYY1551 pKa = 10.6TYY1553 pKa = 11.62DD1554 pKa = 3.42VTNTGNLTITDD1565 pKa = 5.9PITVSDD1571 pKa = 4.4DD1572 pKa = 3.58KK1573 pKa = 11.08IATVNCPALPSGGLAPGAMLTCTASYY1599 pKa = 9.8TITQADD1605 pKa = 3.37VDD1607 pKa = 4.25AGSVTNLATATSGTVNSNQATEE1629 pKa = 4.4TVNATRR1635 pKa = 11.84SPSLGLEE1642 pKa = 4.18KK1643 pKa = 9.88TASPLVYY1650 pKa = 10.05AAAGDD1655 pKa = 4.15VISYY1659 pKa = 10.61SYY1661 pKa = 11.63LLTNTGNVTLSAPYY1675 pKa = 9.2TVADD1679 pKa = 4.41DD1680 pKa = 4.2KK1681 pKa = 10.68TTVTCPATPGTLAPGGTVTCTATYY1705 pKa = 9.36TITQTDD1711 pKa = 3.69VNAGSVTNVATASAIFNNAAVNSAQDD1737 pKa = 3.58SATVTATQTPTLSLDD1752 pKa = 3.06KK1753 pKa = 10.63RR1754 pKa = 11.84LVSGAGFDD1762 pKa = 3.74SVGDD1766 pKa = 4.05VIQYY1770 pKa = 10.88AYY1772 pKa = 10.84DD1773 pKa = 3.47VTNSGNVTITGPITISDD1790 pKa = 3.9DD1791 pKa = 3.72KK1792 pKa = 10.6IASVTCPAGDD1802 pKa = 4.08LAPGAMVTCTASYY1815 pKa = 9.77TVTQADD1821 pKa = 3.93INTGSVTNIATAAGSFNNQPVTSNPDD1847 pKa = 3.38TVTVEE1852 pKa = 4.06ALQEE1856 pKa = 3.87PALTLDD1862 pKa = 3.41KK1863 pKa = 10.69RR1864 pKa = 11.84VVSGNPYY1871 pKa = 10.64DD1872 pKa = 3.65SVGDD1876 pKa = 3.93VVQYY1880 pKa = 10.6TYY1882 pKa = 11.62DD1883 pKa = 3.42VTNTGNLTITDD1894 pKa = 5.9PITVSDD1900 pKa = 4.4DD1901 pKa = 3.58KK1902 pKa = 11.08IATVNCPALPSGGLAPGAMLTCTASYY1928 pKa = 9.84TITQADD1934 pKa = 3.82INNGSVTNVATASSGTTQSPPDD1956 pKa = 4.09TVTVTATQRR1965 pKa = 11.84PALSLDD1971 pKa = 2.97KK1972 pKa = 10.66RR1973 pKa = 11.84LLSGTTVTAVGDD1985 pKa = 4.18VIQYY1989 pKa = 10.88AYY1991 pKa = 10.66DD1992 pKa = 3.38VRR1994 pKa = 11.84NTGNVTITGPITVSDD2009 pKa = 4.08DD2010 pKa = 3.64KK2011 pKa = 11.05IATVTCPAGDD2021 pKa = 4.1LAPAAMLTCTASYY2034 pKa = 9.83TVTQADD2040 pKa = 4.31LDD2042 pKa = 4.2AGSITNIATASGTFNEE2058 pKa = 4.34QTVTSNQDD2066 pKa = 3.09SVTVEE2071 pKa = 4.36ALHH2074 pKa = 6.21EE2075 pKa = 4.14MAVVLTKK2082 pKa = 10.23IAAKK2086 pKa = 10.32RR2087 pKa = 11.84EE2088 pKa = 3.99VRR2090 pKa = 11.84VGDD2093 pKa = 3.5LVRR2096 pKa = 11.84FTVVARR2102 pKa = 11.84NVGPGPVVDD2111 pKa = 4.23ATLIDD2116 pKa = 3.69TPPRR2120 pKa = 11.84GFTFVEE2126 pKa = 5.03GSLTADD2132 pKa = 3.93DD2133 pKa = 4.69EE2134 pKa = 4.99DD2135 pKa = 5.89DD2136 pKa = 3.81NVVVSGVDD2144 pKa = 3.7PIRR2147 pKa = 11.84ISGVDD2152 pKa = 2.82IATNGTATFVYY2163 pKa = 9.6FLRR2166 pKa = 11.84VGAGVGHH2173 pKa = 6.22GTHH2176 pKa = 6.85INRR2179 pKa = 11.84VIAVDD2184 pKa = 3.75PTEE2187 pKa = 4.06HH2188 pKa = 5.81TVSNEE2193 pKa = 3.11AAAEE2197 pKa = 4.1VVIIGDD2203 pKa = 3.69PMLDD2207 pKa = 3.1EE2208 pKa = 4.38SLIVGTVFDD2217 pKa = 5.08DD2218 pKa = 4.0RR2219 pKa = 11.84NGDD2222 pKa = 4.27GIQQPGEE2229 pKa = 3.78RR2230 pKa = 11.84GIPGVRR2236 pKa = 11.84IGSVEE2241 pKa = 3.96GLIMEE2246 pKa = 4.67TDD2248 pKa = 2.98AFGRR2252 pKa = 11.84YY2253 pKa = 8.87HH2254 pKa = 7.17LVGIDD2259 pKa = 3.19GGDD2262 pKa = 3.82FSRR2265 pKa = 11.84GRR2267 pKa = 11.84NFILKK2272 pKa = 9.58VDD2274 pKa = 3.96ASTLPVGTEE2283 pKa = 3.9FTTEE2287 pKa = 3.55NPRR2290 pKa = 11.84VRR2292 pKa = 11.84RR2293 pKa = 11.84ITPGVPVRR2301 pKa = 11.84FDD2303 pKa = 3.65FGVKK2307 pKa = 10.25LPNGEE2312 pKa = 4.21MKK2314 pKa = 10.61GGRR2317 pKa = 11.84QEE2319 pKa = 4.53LDD2321 pKa = 3.2AEE2323 pKa = 4.48LGEE2326 pKa = 4.58VMFDD2330 pKa = 3.63PRR2332 pKa = 11.84SSTLKK2337 pKa = 10.78AEE2339 pKa = 4.1YY2340 pKa = 10.44LPVIDD2345 pKa = 5.05KK2346 pKa = 10.39VASKK2350 pKa = 10.37VRR2352 pKa = 11.84EE2353 pKa = 4.09YY2354 pKa = 11.14NGGRR2358 pKa = 11.84IVITAKK2364 pKa = 10.81AEE2366 pKa = 4.06EE2367 pKa = 4.19EE2368 pKa = 3.88ALAYY2372 pKa = 10.31ARR2374 pKa = 11.84ARR2376 pKa = 11.84AVQDD2380 pKa = 3.36ALRR2383 pKa = 11.84TKK2385 pKa = 10.76LEE2387 pKa = 3.91PALADD2392 pKa = 3.56RR2393 pKa = 11.84VAVEE2397 pKa = 4.45VVADD2401 pKa = 3.71TSGTMVRR2408 pKa = 11.84LDD2410 pKa = 3.55RR2411 pKa = 11.84GIHH2414 pKa = 6.1LGTFLFDD2421 pKa = 3.48TDD2423 pKa = 3.56KK2424 pKa = 11.54SVIKK2428 pKa = 9.46PQYY2431 pKa = 9.14RR2432 pKa = 11.84GLIKK2436 pKa = 10.57EE2437 pKa = 4.18IADD2440 pKa = 3.52DD2441 pKa = 4.31LNRR2444 pKa = 11.84MGRR2447 pKa = 11.84GTIGLVGHH2455 pKa = 7.23ADD2457 pKa = 3.06MRR2459 pKa = 11.84ASNEE2463 pKa = 3.67YY2464 pKa = 10.28NRR2466 pKa = 11.84YY2467 pKa = 9.65LGLRR2471 pKa = 11.84RR2472 pKa = 11.84SKK2474 pKa = 11.19AVFDD2478 pKa = 5.45AILAQLRR2485 pKa = 11.84PEE2487 pKa = 3.76VRR2489 pKa = 11.84QRR2491 pKa = 11.84VRR2493 pKa = 11.84VDD2495 pKa = 2.73ISDD2498 pKa = 4.45EE2499 pKa = 3.86IDD2501 pKa = 3.36AEE2503 pKa = 4.42TGVGDD2508 pKa = 3.73RR2509 pKa = 4.83
Molecular weight: 255.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0F0J3|A0A0A0F0J3_9GAMM 4Fe-4S ferredoxin-type domain-containing protein OS=Lysobacter daejeonensis GH1-9 OX=1385517 GN=N800_10895 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.31ALCHH6 pKa = 6.03VIAVLVMGLPTPAWSQQAGEE26 pKa = 4.15LRR28 pKa = 11.84KK29 pKa = 9.96CVSPGGAVSFQQQPCAAGSRR49 pKa = 11.84QTSSRR54 pKa = 11.84SYY56 pKa = 9.39VAEE59 pKa = 4.09PAPTAEE65 pKa = 4.43QIRR68 pKa = 11.84ARR70 pKa = 11.84ATRR73 pKa = 11.84EE74 pKa = 3.42QVARR78 pKa = 11.84AEE80 pKa = 4.22SAEE83 pKa = 3.93LSRR86 pKa = 11.84RR87 pKa = 11.84AGTSGHH93 pKa = 6.91LSAPPGRR100 pKa = 11.84GTLHH104 pKa = 6.39RR105 pKa = 11.84VAIAKK110 pKa = 10.09DD111 pKa = 3.34DD112 pKa = 4.21AACQRR117 pKa = 11.84ARR119 pKa = 11.84RR120 pKa = 11.84HH121 pKa = 5.79RR122 pKa = 11.84DD123 pKa = 2.89EE124 pKa = 3.87TLEE127 pKa = 3.95RR128 pKa = 11.84VGLKK132 pKa = 8.79RR133 pKa = 11.84TYY135 pKa = 11.23DD136 pKa = 3.66LLRR139 pKa = 11.84ALNDD143 pKa = 3.32EE144 pKa = 4.39VARR147 pKa = 11.84ACRR150 pKa = 3.43
MM1 pKa = 7.62KK2 pKa = 10.31ALCHH6 pKa = 6.03VIAVLVMGLPTPAWSQQAGEE26 pKa = 4.15LRR28 pKa = 11.84KK29 pKa = 9.96CVSPGGAVSFQQQPCAAGSRR49 pKa = 11.84QTSSRR54 pKa = 11.84SYY56 pKa = 9.39VAEE59 pKa = 4.09PAPTAEE65 pKa = 4.43QIRR68 pKa = 11.84ARR70 pKa = 11.84ATRR73 pKa = 11.84EE74 pKa = 3.42QVARR78 pKa = 11.84AEE80 pKa = 4.22SAEE83 pKa = 3.93LSRR86 pKa = 11.84RR87 pKa = 11.84AGTSGHH93 pKa = 6.91LSAPPGRR100 pKa = 11.84GTLHH104 pKa = 6.39RR105 pKa = 11.84VAIAKK110 pKa = 10.09DD111 pKa = 3.34DD112 pKa = 4.21AACQRR117 pKa = 11.84ARR119 pKa = 11.84RR120 pKa = 11.84HH121 pKa = 5.79RR122 pKa = 11.84DD123 pKa = 2.89EE124 pKa = 3.87TLEE127 pKa = 3.95RR128 pKa = 11.84VGLKK132 pKa = 8.79RR133 pKa = 11.84TYY135 pKa = 11.23DD136 pKa = 3.66LLRR139 pKa = 11.84ALNDD143 pKa = 3.32EE144 pKa = 4.39VARR147 pKa = 11.84ACRR150 pKa = 3.43
Molecular weight: 16.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
830463 |
41 |
2509 |
323.1 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.135 ± 0.064 | 0.825 ± 0.015 |
5.829 ± 0.04 | 5.474 ± 0.04 |
3.376 ± 0.031 | 8.628 ± 0.046 |
2.341 ± 0.026 | 4.044 ± 0.035 |
2.932 ± 0.036 | 10.688 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.266 ± 0.022 | 2.457 ± 0.035 |
5.271 ± 0.033 | 3.543 ± 0.028 |
7.554 ± 0.054 | 4.873 ± 0.037 |
5.082 ± 0.047 | 7.844 ± 0.043 |
1.505 ± 0.027 | 2.332 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
