
Amazon lily mild mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Anulavirus
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
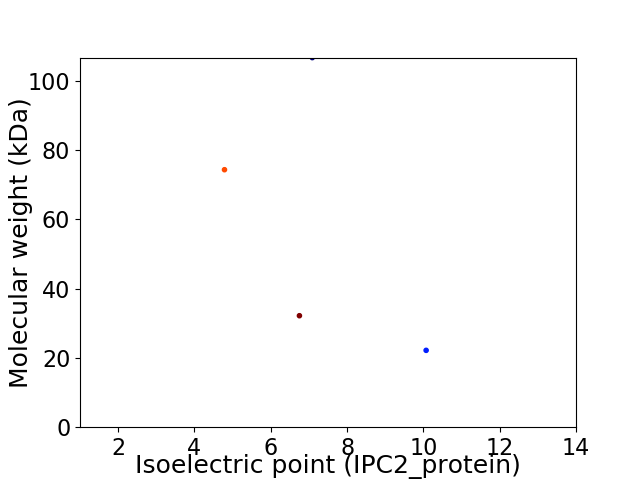
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I7HHJ1|I7HHJ1_9BROM RNA-directed RNA polymerase 2a OS=Amazon lily mild mottle virus OX=1195163 GN=2a PE=4 SV=1
MM1 pKa = 7.87ADD3 pKa = 3.84CLVDD7 pKa = 4.77LGIPKK12 pKa = 10.3LKK14 pKa = 10.46LVAALFRR21 pKa = 11.84GEE23 pKa = 4.95DD24 pKa = 3.26ISDD27 pKa = 3.61ISPEE31 pKa = 4.25VVTEE35 pKa = 4.07VCDD38 pKa = 4.17GSFVSYY44 pKa = 10.54HH45 pKa = 6.42CLDD48 pKa = 3.89SSVSPFDD55 pKa = 3.39VCVDD59 pKa = 3.09ILDD62 pKa = 4.4TIKK65 pKa = 11.14NDD67 pKa = 3.09VSVVDD72 pKa = 4.08DD73 pKa = 4.19CVVTTSDD80 pKa = 3.89EE81 pKa = 4.41EE82 pKa = 4.54VTSEE86 pKa = 3.53RR87 pKa = 11.84LEE89 pKa = 4.13RR90 pKa = 11.84VKK92 pKa = 10.37RR93 pKa = 11.84TLDD96 pKa = 4.08LEE98 pKa = 4.56DD99 pKa = 4.52CVVNSCSVCTEE110 pKa = 3.82GNEE113 pKa = 4.01RR114 pKa = 11.84LMEE117 pKa = 4.24SSYY120 pKa = 11.67GNHH123 pKa = 5.25FTLVGEE129 pKa = 4.42EE130 pKa = 4.59FIEE133 pKa = 4.39EE134 pKa = 4.15CPAAAINVASVVPIDD149 pKa = 3.74QQFEE153 pKa = 4.23CDD155 pKa = 3.52LTAPLPVLKK164 pKa = 10.51GRR166 pKa = 11.84SDD168 pKa = 3.28RR169 pKa = 11.84VTLEE173 pKa = 3.88KK174 pKa = 10.93LQTTVDD180 pKa = 4.25AILPTHH186 pKa = 6.57ARR188 pKa = 11.84FDD190 pKa = 4.07DD191 pKa = 3.97TFHH194 pKa = 6.55QAFVEE199 pKa = 4.18NADD202 pKa = 3.29VSLDD206 pKa = 3.54FEE208 pKa = 5.03RR209 pKa = 11.84IRR211 pKa = 11.84LKK213 pKa = 10.67QYY215 pKa = 10.64AGDD218 pKa = 3.82WYY220 pKa = 10.79RR221 pKa = 11.84DD222 pKa = 3.17PDD224 pKa = 3.69NFYY227 pKa = 11.41EE228 pKa = 4.36PVLKK232 pKa = 10.73SGGCSRR238 pKa = 11.84RR239 pKa = 11.84VGSQKK244 pKa = 10.28EE245 pKa = 3.65ALIAIRR251 pKa = 11.84KK252 pKa = 9.04RR253 pKa = 11.84NADD256 pKa = 3.5VPEE259 pKa = 4.17LAGSVNIDD267 pKa = 4.26QVAEE271 pKa = 3.99WAADD275 pKa = 3.26NFMKK279 pKa = 10.74SFVVNVSPLVQVMSKK294 pKa = 9.08MKK296 pKa = 10.45AYY298 pKa = 9.05MEE300 pKa = 4.2KK301 pKa = 9.88WSDD304 pKa = 3.4RR305 pKa = 11.84VDD307 pKa = 3.31PLMVLGEE314 pKa = 4.5TNLQRR319 pKa = 11.84YY320 pKa = 6.28QHH322 pKa = 6.19MIKK325 pKa = 9.6TDD327 pKa = 3.42VKK329 pKa = 10.65PITNDD334 pKa = 2.75GMNLEE339 pKa = 4.26RR340 pKa = 11.84AIPATITFHH349 pKa = 7.85DD350 pKa = 4.74KK351 pKa = 9.86MVCSNFSPWFTALFDD366 pKa = 4.24GLQKK370 pKa = 10.84SLNSRR375 pKa = 11.84VRR377 pKa = 11.84IPSGPISTLEE387 pKa = 3.45MHH389 pKa = 6.68YY390 pKa = 10.87GFKK393 pKa = 10.47NKK395 pKa = 10.36YY396 pKa = 8.25YY397 pKa = 11.06VEE399 pKa = 4.79IDD401 pKa = 3.19LSKK404 pKa = 10.49FDD406 pKa = 3.77KK407 pKa = 11.17SQGEE411 pKa = 3.98LHH413 pKa = 7.1LEE415 pKa = 4.14FQRR418 pKa = 11.84LILLRR423 pKa = 11.84LGLPAHH429 pKa = 6.75LVNWWCEE436 pKa = 3.33LHH438 pKa = 6.31IRR440 pKa = 11.84SFISDD445 pKa = 3.66PTAGVAFQCAYY456 pKa = 9.14QRR458 pKa = 11.84RR459 pKa = 11.84TGDD462 pKa = 2.78AFTFFGNTLVTMAEE476 pKa = 4.43FACCFDD482 pKa = 3.99CSQFEE487 pKa = 4.24MMLFAGDD494 pKa = 3.89DD495 pKa = 3.69SLAVSNSPITGDD507 pKa = 3.17TDD509 pKa = 4.61LFTSLFNMEE518 pKa = 4.88SKK520 pKa = 11.15SMANPVPYY528 pKa = 9.81ICSKK532 pKa = 10.61FLIEE536 pKa = 4.54DD537 pKa = 3.57CFGNSFSVPDD547 pKa = 5.15PIRR550 pKa = 11.84EE551 pKa = 3.88FQRR554 pKa = 11.84LGKK557 pKa = 10.23KK558 pKa = 9.89KK559 pKa = 10.16IQIQPRR565 pKa = 11.84ADD567 pKa = 3.26ALFEE571 pKa = 4.06QYY573 pKa = 10.54QGFRR577 pKa = 11.84DD578 pKa = 3.29RR579 pKa = 11.84MKK581 pKa = 10.75YY582 pKa = 10.0LRR584 pKa = 11.84HH585 pKa = 6.59LDD587 pKa = 3.86DD588 pKa = 6.15FMIDD592 pKa = 3.32QLKK595 pKa = 10.35VYY597 pKa = 10.72FDD599 pKa = 3.24IKK601 pKa = 10.09YY602 pKa = 10.85NKK604 pKa = 10.38GKK606 pKa = 10.44DD607 pKa = 3.79CIDD610 pKa = 4.36DD611 pKa = 3.79FLGGCIYY618 pKa = 10.85YY619 pKa = 10.64ADD621 pKa = 3.97NFKK624 pKa = 10.69HH625 pKa = 5.56FCEE628 pKa = 4.37LFVNHH633 pKa = 5.68SHH635 pKa = 6.96AIEE638 pKa = 4.14VGALVNFVDD647 pKa = 5.31DD648 pKa = 4.67KK649 pKa = 11.42PLGIPSRR656 pKa = 11.84LL657 pKa = 3.38
MM1 pKa = 7.87ADD3 pKa = 3.84CLVDD7 pKa = 4.77LGIPKK12 pKa = 10.3LKK14 pKa = 10.46LVAALFRR21 pKa = 11.84GEE23 pKa = 4.95DD24 pKa = 3.26ISDD27 pKa = 3.61ISPEE31 pKa = 4.25VVTEE35 pKa = 4.07VCDD38 pKa = 4.17GSFVSYY44 pKa = 10.54HH45 pKa = 6.42CLDD48 pKa = 3.89SSVSPFDD55 pKa = 3.39VCVDD59 pKa = 3.09ILDD62 pKa = 4.4TIKK65 pKa = 11.14NDD67 pKa = 3.09VSVVDD72 pKa = 4.08DD73 pKa = 4.19CVVTTSDD80 pKa = 3.89EE81 pKa = 4.41EE82 pKa = 4.54VTSEE86 pKa = 3.53RR87 pKa = 11.84LEE89 pKa = 4.13RR90 pKa = 11.84VKK92 pKa = 10.37RR93 pKa = 11.84TLDD96 pKa = 4.08LEE98 pKa = 4.56DD99 pKa = 4.52CVVNSCSVCTEE110 pKa = 3.82GNEE113 pKa = 4.01RR114 pKa = 11.84LMEE117 pKa = 4.24SSYY120 pKa = 11.67GNHH123 pKa = 5.25FTLVGEE129 pKa = 4.42EE130 pKa = 4.59FIEE133 pKa = 4.39EE134 pKa = 4.15CPAAAINVASVVPIDD149 pKa = 3.74QQFEE153 pKa = 4.23CDD155 pKa = 3.52LTAPLPVLKK164 pKa = 10.51GRR166 pKa = 11.84SDD168 pKa = 3.28RR169 pKa = 11.84VTLEE173 pKa = 3.88KK174 pKa = 10.93LQTTVDD180 pKa = 4.25AILPTHH186 pKa = 6.57ARR188 pKa = 11.84FDD190 pKa = 4.07DD191 pKa = 3.97TFHH194 pKa = 6.55QAFVEE199 pKa = 4.18NADD202 pKa = 3.29VSLDD206 pKa = 3.54FEE208 pKa = 5.03RR209 pKa = 11.84IRR211 pKa = 11.84LKK213 pKa = 10.67QYY215 pKa = 10.64AGDD218 pKa = 3.82WYY220 pKa = 10.79RR221 pKa = 11.84DD222 pKa = 3.17PDD224 pKa = 3.69NFYY227 pKa = 11.41EE228 pKa = 4.36PVLKK232 pKa = 10.73SGGCSRR238 pKa = 11.84RR239 pKa = 11.84VGSQKK244 pKa = 10.28EE245 pKa = 3.65ALIAIRR251 pKa = 11.84KK252 pKa = 9.04RR253 pKa = 11.84NADD256 pKa = 3.5VPEE259 pKa = 4.17LAGSVNIDD267 pKa = 4.26QVAEE271 pKa = 3.99WAADD275 pKa = 3.26NFMKK279 pKa = 10.74SFVVNVSPLVQVMSKK294 pKa = 9.08MKK296 pKa = 10.45AYY298 pKa = 9.05MEE300 pKa = 4.2KK301 pKa = 9.88WSDD304 pKa = 3.4RR305 pKa = 11.84VDD307 pKa = 3.31PLMVLGEE314 pKa = 4.5TNLQRR319 pKa = 11.84YY320 pKa = 6.28QHH322 pKa = 6.19MIKK325 pKa = 9.6TDD327 pKa = 3.42VKK329 pKa = 10.65PITNDD334 pKa = 2.75GMNLEE339 pKa = 4.26RR340 pKa = 11.84AIPATITFHH349 pKa = 7.85DD350 pKa = 4.74KK351 pKa = 9.86MVCSNFSPWFTALFDD366 pKa = 4.24GLQKK370 pKa = 10.84SLNSRR375 pKa = 11.84VRR377 pKa = 11.84IPSGPISTLEE387 pKa = 3.45MHH389 pKa = 6.68YY390 pKa = 10.87GFKK393 pKa = 10.47NKK395 pKa = 10.36YY396 pKa = 8.25YY397 pKa = 11.06VEE399 pKa = 4.79IDD401 pKa = 3.19LSKK404 pKa = 10.49FDD406 pKa = 3.77KK407 pKa = 11.17SQGEE411 pKa = 3.98LHH413 pKa = 7.1LEE415 pKa = 4.14FQRR418 pKa = 11.84LILLRR423 pKa = 11.84LGLPAHH429 pKa = 6.75LVNWWCEE436 pKa = 3.33LHH438 pKa = 6.31IRR440 pKa = 11.84SFISDD445 pKa = 3.66PTAGVAFQCAYY456 pKa = 9.14QRR458 pKa = 11.84RR459 pKa = 11.84TGDD462 pKa = 2.78AFTFFGNTLVTMAEE476 pKa = 4.43FACCFDD482 pKa = 3.99CSQFEE487 pKa = 4.24MMLFAGDD494 pKa = 3.89DD495 pKa = 3.69SLAVSNSPITGDD507 pKa = 3.17TDD509 pKa = 4.61LFTSLFNMEE518 pKa = 4.88SKK520 pKa = 11.15SMANPVPYY528 pKa = 9.81ICSKK532 pKa = 10.61FLIEE536 pKa = 4.54DD537 pKa = 3.57CFGNSFSVPDD547 pKa = 5.15PIRR550 pKa = 11.84EE551 pKa = 3.88FQRR554 pKa = 11.84LGKK557 pKa = 10.23KK558 pKa = 9.89KK559 pKa = 10.16IQIQPRR565 pKa = 11.84ADD567 pKa = 3.26ALFEE571 pKa = 4.06QYY573 pKa = 10.54QGFRR577 pKa = 11.84DD578 pKa = 3.29RR579 pKa = 11.84MKK581 pKa = 10.75YY582 pKa = 10.0LRR584 pKa = 11.84HH585 pKa = 6.59LDD587 pKa = 3.86DD588 pKa = 6.15FMIDD592 pKa = 3.32QLKK595 pKa = 10.35VYY597 pKa = 10.72FDD599 pKa = 3.24IKK601 pKa = 10.09YY602 pKa = 10.85NKK604 pKa = 10.38GKK606 pKa = 10.44DD607 pKa = 3.79CIDD610 pKa = 4.36DD611 pKa = 3.79FLGGCIYY618 pKa = 10.85YY619 pKa = 10.64ADD621 pKa = 3.97NFKK624 pKa = 10.69HH625 pKa = 5.56FCEE628 pKa = 4.37LFVNHH633 pKa = 5.68SHH635 pKa = 6.96AIEE638 pKa = 4.14VGALVNFVDD647 pKa = 5.31DD648 pKa = 4.67KK649 pKa = 11.42PLGIPSRR656 pKa = 11.84LL657 pKa = 3.38
Molecular weight: 74.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I7H7W8|I7H7W8_9BROM Movement protein OS=Amazon lily mild mottle virus OX=1195163 GN=3a PE=4 SV=1
MM1 pKa = 7.24QNPNRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84NRR10 pKa = 11.84ANQNRR15 pKa = 11.84MRR17 pKa = 11.84RR18 pKa = 11.84QRR20 pKa = 11.84QAQDD24 pKa = 2.78AAAFRR29 pKa = 11.84ALSSSSRR36 pKa = 11.84LGPSTSALVDD46 pKa = 3.43AGGIPVTPGFVVTSVTLSEE65 pKa = 4.1AFQWSGPGTGAWTLFRR81 pKa = 11.84EE82 pKa = 4.32RR83 pKa = 11.84SLVIPSALPPEE94 pKa = 4.28TRR96 pKa = 11.84LNNVILRR103 pKa = 11.84VTPTPGLASFDD114 pKa = 2.91IWVAVAAAKK123 pKa = 10.52GSTPTAVDD131 pKa = 4.28FDD133 pKa = 4.28ALNVPFLTMRR143 pKa = 11.84EE144 pKa = 4.07TSRR147 pKa = 11.84TRR149 pKa = 11.84HH150 pKa = 4.14VVVPFVNRR158 pKa = 11.84TVADD162 pKa = 4.45LAQQRR167 pKa = 11.84VWLGARR173 pKa = 11.84TGTVTAEE180 pKa = 4.03SLVVGLVRR188 pKa = 11.84VFIEE192 pKa = 4.36HH193 pKa = 6.84RR194 pKa = 11.84PIPQVQIVPVV204 pKa = 3.48
MM1 pKa = 7.24QNPNRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84NRR10 pKa = 11.84ANQNRR15 pKa = 11.84MRR17 pKa = 11.84RR18 pKa = 11.84QRR20 pKa = 11.84QAQDD24 pKa = 2.78AAAFRR29 pKa = 11.84ALSSSSRR36 pKa = 11.84LGPSTSALVDD46 pKa = 3.43AGGIPVTPGFVVTSVTLSEE65 pKa = 4.1AFQWSGPGTGAWTLFRR81 pKa = 11.84EE82 pKa = 4.32RR83 pKa = 11.84SLVIPSALPPEE94 pKa = 4.28TRR96 pKa = 11.84LNNVILRR103 pKa = 11.84VTPTPGLASFDD114 pKa = 2.91IWVAVAAAKK123 pKa = 10.52GSTPTAVDD131 pKa = 4.28FDD133 pKa = 4.28ALNVPFLTMRR143 pKa = 11.84EE144 pKa = 4.07TSRR147 pKa = 11.84TRR149 pKa = 11.84HH150 pKa = 4.14VVVPFVNRR158 pKa = 11.84TVADD162 pKa = 4.45LAQQRR167 pKa = 11.84VWLGARR173 pKa = 11.84TGTVTAEE180 pKa = 4.03SLVVGLVRR188 pKa = 11.84VFIEE192 pKa = 4.36HH193 pKa = 6.84RR194 pKa = 11.84PIPQVQIVPVV204 pKa = 3.48
Molecular weight: 22.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2102 |
204 |
945 |
525.5 |
58.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.279 ± 0.736 | 2.854 ± 0.56 |
6.708 ± 0.908 | 5.852 ± 0.667 |
5.186 ± 0.59 | 5.519 ± 0.489 |
2.093 ± 0.443 | 4.567 ± 0.749 |
6.185 ± 1.301 | 9.324 ± 0.497 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.216 | 3.806 ± 0.197 |
4.044 ± 0.982 | 2.95 ± 0.396 |
5.709 ± 0.809 | 7.85 ± 1.008 |
5.233 ± 0.596 | 8.706 ± 0.695 |
1.38 ± 0.191 | 2.521 ± 0.414 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
