
Cuiaba virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sripuvirus; Cuiaba sripuvirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
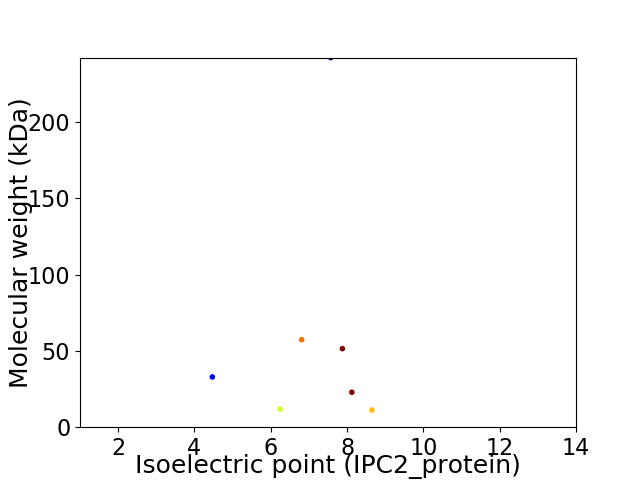
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8TMP8|A0A3S8TMP8_9RHAB U1 OS=Cuiaba virus OX=2495751 GN=U1 PE=4 SV=1
MM1 pKa = 7.38MSSQQSFPKK10 pKa = 10.7DD11 pKa = 3.33FDD13 pKa = 3.63PTKK16 pKa = 9.98TLEE19 pKa = 3.83AAKK22 pKa = 10.37RR23 pKa = 11.84GLKK26 pKa = 10.3KK27 pKa = 10.77LEE29 pKa = 4.6SDD31 pKa = 3.9DD32 pKa = 4.48KK33 pKa = 11.48MMFGVEE39 pKa = 4.12SLDD42 pKa = 4.44DD43 pKa = 4.78EE44 pKa = 4.66IEE46 pKa = 4.02KK47 pKa = 10.23HH48 pKa = 5.3QEE50 pKa = 3.74SSSDD54 pKa = 3.47EE55 pKa = 4.24EE56 pKa = 4.38EE57 pKa = 5.11EE58 pKa = 4.67EE59 pKa = 4.14EE60 pKa = 5.42CDD62 pKa = 4.47DD63 pKa = 4.1NVSKK67 pKa = 10.78EE68 pKa = 4.02AVPDD72 pKa = 3.97KK73 pKa = 10.74EE74 pKa = 4.13QLEE77 pKa = 4.22ADD79 pKa = 3.97DD80 pKa = 5.45FLEE83 pKa = 5.07QEE85 pKa = 4.58SSWTFNDD92 pKa = 2.91SKK94 pKa = 11.53LYY96 pKa = 7.45ITKK99 pKa = 10.08HH100 pKa = 5.0FSVNKK105 pKa = 9.31DD106 pKa = 2.94TMDD109 pKa = 3.81LVNAHH114 pKa = 6.32IRR116 pKa = 11.84NSVEE120 pKa = 3.49KK121 pKa = 9.64TLNFLGAKK129 pKa = 8.1ATYY132 pKa = 9.37EE133 pKa = 4.24GEE135 pKa = 4.54DD136 pKa = 3.56GSSFAWRR143 pKa = 11.84VSSDD147 pKa = 3.74AIHH150 pKa = 6.69PMEE153 pKa = 4.63KK154 pKa = 9.72HH155 pKa = 5.59YY156 pKa = 10.96VWQEE160 pKa = 3.47EE161 pKa = 4.34TQRR164 pKa = 11.84PIPSAPPSYY173 pKa = 10.86EE174 pKa = 4.77DD175 pKa = 4.56LDD177 pKa = 4.05LPSRR181 pKa = 11.84GKK183 pKa = 9.03TSPPLRR189 pKa = 11.84TSEE192 pKa = 4.16DD193 pKa = 3.39TTKK196 pKa = 10.7PDD198 pKa = 3.33YY199 pKa = 10.55QKK201 pKa = 10.8PSTSSEE207 pKa = 3.83CRR209 pKa = 11.84KK210 pKa = 10.24QSDD213 pKa = 3.48LSINRR218 pKa = 11.84EE219 pKa = 3.82FLDD222 pKa = 3.67SFKK225 pKa = 11.04KK226 pKa = 10.85GILFPSIDD234 pKa = 3.68GGGDD238 pKa = 3.18ILVKK242 pKa = 10.7LADD245 pKa = 3.76TDD247 pKa = 3.69LTEE250 pKa = 5.17AIIRR254 pKa = 11.84DD255 pKa = 4.04FVYY258 pKa = 10.61EE259 pKa = 4.33PSTSYY264 pKa = 11.39QDD266 pKa = 2.91MVYY269 pKa = 10.3QIFCIHH275 pKa = 6.95PQTKK279 pKa = 9.23PDD281 pKa = 3.43YY282 pKa = 11.1EE283 pKa = 4.54FIRR286 pKa = 11.84WW287 pKa = 3.37
MM1 pKa = 7.38MSSQQSFPKK10 pKa = 10.7DD11 pKa = 3.33FDD13 pKa = 3.63PTKK16 pKa = 9.98TLEE19 pKa = 3.83AAKK22 pKa = 10.37RR23 pKa = 11.84GLKK26 pKa = 10.3KK27 pKa = 10.77LEE29 pKa = 4.6SDD31 pKa = 3.9DD32 pKa = 4.48KK33 pKa = 11.48MMFGVEE39 pKa = 4.12SLDD42 pKa = 4.44DD43 pKa = 4.78EE44 pKa = 4.66IEE46 pKa = 4.02KK47 pKa = 10.23HH48 pKa = 5.3QEE50 pKa = 3.74SSSDD54 pKa = 3.47EE55 pKa = 4.24EE56 pKa = 4.38EE57 pKa = 5.11EE58 pKa = 4.67EE59 pKa = 4.14EE60 pKa = 5.42CDD62 pKa = 4.47DD63 pKa = 4.1NVSKK67 pKa = 10.78EE68 pKa = 4.02AVPDD72 pKa = 3.97KK73 pKa = 10.74EE74 pKa = 4.13QLEE77 pKa = 4.22ADD79 pKa = 3.97DD80 pKa = 5.45FLEE83 pKa = 5.07QEE85 pKa = 4.58SSWTFNDD92 pKa = 2.91SKK94 pKa = 11.53LYY96 pKa = 7.45ITKK99 pKa = 10.08HH100 pKa = 5.0FSVNKK105 pKa = 9.31DD106 pKa = 2.94TMDD109 pKa = 3.81LVNAHH114 pKa = 6.32IRR116 pKa = 11.84NSVEE120 pKa = 3.49KK121 pKa = 9.64TLNFLGAKK129 pKa = 8.1ATYY132 pKa = 9.37EE133 pKa = 4.24GEE135 pKa = 4.54DD136 pKa = 3.56GSSFAWRR143 pKa = 11.84VSSDD147 pKa = 3.74AIHH150 pKa = 6.69PMEE153 pKa = 4.63KK154 pKa = 9.72HH155 pKa = 5.59YY156 pKa = 10.96VWQEE160 pKa = 3.47EE161 pKa = 4.34TQRR164 pKa = 11.84PIPSAPPSYY173 pKa = 10.86EE174 pKa = 4.77DD175 pKa = 4.56LDD177 pKa = 4.05LPSRR181 pKa = 11.84GKK183 pKa = 9.03TSPPLRR189 pKa = 11.84TSEE192 pKa = 4.16DD193 pKa = 3.39TTKK196 pKa = 10.7PDD198 pKa = 3.33YY199 pKa = 10.55QKK201 pKa = 10.8PSTSSEE207 pKa = 3.83CRR209 pKa = 11.84KK210 pKa = 10.24QSDD213 pKa = 3.48LSINRR218 pKa = 11.84EE219 pKa = 3.82FLDD222 pKa = 3.67SFKK225 pKa = 11.04KK226 pKa = 10.85GILFPSIDD234 pKa = 3.68GGGDD238 pKa = 3.18ILVKK242 pKa = 10.7LADD245 pKa = 3.76TDD247 pKa = 3.69LTEE250 pKa = 5.17AIIRR254 pKa = 11.84DD255 pKa = 4.04FVYY258 pKa = 10.61EE259 pKa = 4.33PSTSYY264 pKa = 11.39QDD266 pKa = 2.91MVYY269 pKa = 10.3QIFCIHH275 pKa = 6.95PQTKK279 pKa = 9.23PDD281 pKa = 3.43YY282 pKa = 11.1EE283 pKa = 4.54FIRR286 pKa = 11.84WW287 pKa = 3.37
Molecular weight: 32.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8TMQ4|A0A3S8TMQ4_9RHAB RNA-directed RNA polymerase L OS=Cuiaba virus OX=2495751 GN=L PE=3 SV=1
MM1 pKa = 7.75EE2 pKa = 4.53KK3 pKa = 9.68TGQKK7 pKa = 9.45PVSGQLSVQKK17 pKa = 10.87LIDD20 pKa = 3.4QGKK23 pKa = 7.59WEE25 pKa = 4.89PRR27 pKa = 11.84PPLNPAKK34 pKa = 10.31RR35 pKa = 11.84CTKK38 pKa = 10.33VEE40 pKa = 4.61DD41 pKa = 3.7LSSWPDD47 pKa = 3.14ITSKK51 pKa = 9.21TCSLFKK57 pKa = 10.78HH58 pKa = 6.18PLSFDD63 pKa = 3.16QGPDD67 pKa = 2.84AKK69 pKa = 10.22RR70 pKa = 11.84GKK72 pKa = 10.13KK73 pKa = 9.72ADD75 pKa = 4.45CIQIRR80 pKa = 11.84SPVFVAKK87 pKa = 9.92TIAQGDD93 pKa = 3.92NQLCMIPRR101 pKa = 4.12
MM1 pKa = 7.75EE2 pKa = 4.53KK3 pKa = 9.68TGQKK7 pKa = 9.45PVSGQLSVQKK17 pKa = 10.87LIDD20 pKa = 3.4QGKK23 pKa = 7.59WEE25 pKa = 4.89PRR27 pKa = 11.84PPLNPAKK34 pKa = 10.31RR35 pKa = 11.84CTKK38 pKa = 10.33VEE40 pKa = 4.61DD41 pKa = 3.7LSSWPDD47 pKa = 3.14ITSKK51 pKa = 9.21TCSLFKK57 pKa = 10.78HH58 pKa = 6.18PLSFDD63 pKa = 3.16QGPDD67 pKa = 2.84AKK69 pKa = 10.22RR70 pKa = 11.84GKK72 pKa = 10.13KK73 pKa = 9.72ADD75 pKa = 4.45CIQIRR80 pKa = 11.84SPVFVAKK87 pKa = 9.92TIAQGDD93 pKa = 3.92NQLCMIPRR101 pKa = 4.12
Molecular weight: 11.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3742 |
101 |
2092 |
534.6 |
61.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.581 ± 0.444 | 2.218 ± 0.26 |
5.879 ± 0.816 | 6.708 ± 0.672 |
4.516 ± 0.203 | 5.692 ± 0.411 |
1.978 ± 0.334 | 7.803 ± 0.904 |
7.723 ± 0.584 | 9.246 ± 0.773 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.859 ± 0.258 | 4.784 ± 0.903 |
4.409 ± 0.559 | 3.18 ± 0.379 |
5.184 ± 0.37 | 8.311 ± 0.55 |
5.184 ± 0.542 | 5.158 ± 0.264 |
1.737 ± 0.204 | 3.848 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
