
Farfantepenaeus duorarum circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.78
Get precalculated fractions of proteins
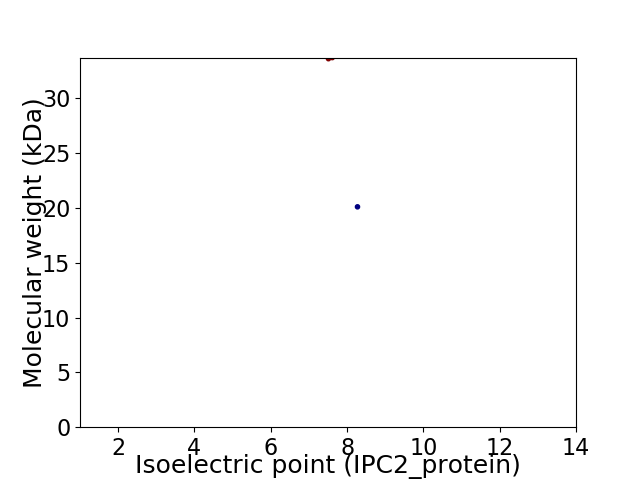
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
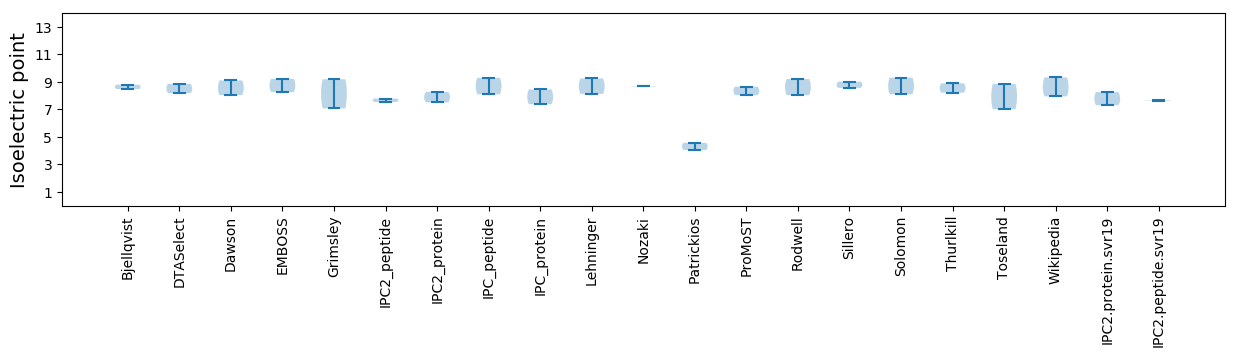
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5U8K1|S5U8K1_9CIRC Capsid protein OS=Farfantepenaeus duorarum circovirus OX=1380894 PE=4 SV=1
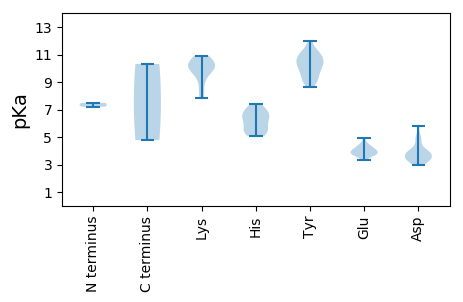
MM1 pKa = 7.21SGSRR5 pKa = 11.84RR6 pKa = 11.84WCFTVFNYY14 pKa = 10.06NHH16 pKa = 6.48ATVDD20 pKa = 3.4DD21 pKa = 5.07FIRR24 pKa = 11.84AEE26 pKa = 3.77RR27 pKa = 11.84HH28 pKa = 5.09DD29 pKa = 3.88CVRR32 pKa = 11.84YY33 pKa = 9.49VVWQCEE39 pKa = 4.09KK40 pKa = 10.49CPTSGRR46 pKa = 11.84SHH48 pKa = 5.56IQGYY52 pKa = 9.08VEE54 pKa = 3.71LHH56 pKa = 5.4RR57 pKa = 11.84PIRR60 pKa = 11.84LRR62 pKa = 11.84QLKK65 pKa = 10.27QILDD69 pKa = 3.65DD70 pKa = 3.94TGAHH74 pKa = 6.24CEE76 pKa = 3.91AAKK79 pKa = 10.43GPRR82 pKa = 11.84DD83 pKa = 3.3ACRR86 pKa = 11.84EE87 pKa = 3.97YY88 pKa = 10.77CQKK91 pKa = 10.85EE92 pKa = 3.99EE93 pKa = 4.49SRR95 pKa = 11.84CCGWWEE101 pKa = 3.84SGEE104 pKa = 4.18WIGGQGSRR112 pKa = 11.84SDD114 pKa = 3.75LSAVHH119 pKa = 6.84AAIQDD124 pKa = 3.86GASLRR129 pKa = 11.84EE130 pKa = 3.72ISNAHH135 pKa = 6.12FDD137 pKa = 3.79AFVKK141 pKa = 9.96YY142 pKa = 10.45SRR144 pKa = 11.84GIKK147 pKa = 9.94EE148 pKa = 4.4YY149 pKa = 10.47ILLNADD155 pKa = 2.95RR156 pKa = 11.84RR157 pKa = 11.84MWPMEE162 pKa = 3.64VWVLYY167 pKa = 11.08GDD169 pKa = 3.89TEE171 pKa = 4.44TGKK174 pKa = 8.17TRR176 pKa = 11.84AVYY179 pKa = 10.43EE180 pKa = 4.15MAPDD184 pKa = 4.21VYY186 pKa = 11.19SLAPSHH192 pKa = 6.8SNGVGWWQMYY202 pKa = 9.24EE203 pKa = 3.72GHH205 pKa = 7.37ADD207 pKa = 3.36VLLDD211 pKa = 5.8DD212 pKa = 4.74FYY214 pKa = 11.96GWLRR218 pKa = 11.84YY219 pKa = 9.92SFLLQLLDD227 pKa = 4.18RR228 pKa = 11.84YY229 pKa = 9.19PLRR232 pKa = 11.84VEE234 pKa = 4.21SKK236 pKa = 9.25GGVVQFVGRR245 pKa = 11.84RR246 pKa = 11.84IFITSNAHH254 pKa = 5.41PNQWYY259 pKa = 9.18PNVKK263 pKa = 8.82DD264 pKa = 3.36TAAMFRR270 pKa = 11.84RR271 pKa = 11.84FTRR274 pKa = 11.84IYY276 pKa = 9.8HH277 pKa = 6.05CDD279 pKa = 3.01KK280 pKa = 10.92SRR282 pKa = 11.84FWRR285 pKa = 11.84EE286 pKa = 3.31KK287 pKa = 10.31
MM1 pKa = 7.21SGSRR5 pKa = 11.84RR6 pKa = 11.84WCFTVFNYY14 pKa = 10.06NHH16 pKa = 6.48ATVDD20 pKa = 3.4DD21 pKa = 5.07FIRR24 pKa = 11.84AEE26 pKa = 3.77RR27 pKa = 11.84HH28 pKa = 5.09DD29 pKa = 3.88CVRR32 pKa = 11.84YY33 pKa = 9.49VVWQCEE39 pKa = 4.09KK40 pKa = 10.49CPTSGRR46 pKa = 11.84SHH48 pKa = 5.56IQGYY52 pKa = 9.08VEE54 pKa = 3.71LHH56 pKa = 5.4RR57 pKa = 11.84PIRR60 pKa = 11.84LRR62 pKa = 11.84QLKK65 pKa = 10.27QILDD69 pKa = 3.65DD70 pKa = 3.94TGAHH74 pKa = 6.24CEE76 pKa = 3.91AAKK79 pKa = 10.43GPRR82 pKa = 11.84DD83 pKa = 3.3ACRR86 pKa = 11.84EE87 pKa = 3.97YY88 pKa = 10.77CQKK91 pKa = 10.85EE92 pKa = 3.99EE93 pKa = 4.49SRR95 pKa = 11.84CCGWWEE101 pKa = 3.84SGEE104 pKa = 4.18WIGGQGSRR112 pKa = 11.84SDD114 pKa = 3.75LSAVHH119 pKa = 6.84AAIQDD124 pKa = 3.86GASLRR129 pKa = 11.84EE130 pKa = 3.72ISNAHH135 pKa = 6.12FDD137 pKa = 3.79AFVKK141 pKa = 9.96YY142 pKa = 10.45SRR144 pKa = 11.84GIKK147 pKa = 9.94EE148 pKa = 4.4YY149 pKa = 10.47ILLNADD155 pKa = 2.95RR156 pKa = 11.84RR157 pKa = 11.84MWPMEE162 pKa = 3.64VWVLYY167 pKa = 11.08GDD169 pKa = 3.89TEE171 pKa = 4.44TGKK174 pKa = 8.17TRR176 pKa = 11.84AVYY179 pKa = 10.43EE180 pKa = 4.15MAPDD184 pKa = 4.21VYY186 pKa = 11.19SLAPSHH192 pKa = 6.8SNGVGWWQMYY202 pKa = 9.24EE203 pKa = 3.72GHH205 pKa = 7.37ADD207 pKa = 3.36VLLDD211 pKa = 5.8DD212 pKa = 4.74FYY214 pKa = 11.96GWLRR218 pKa = 11.84YY219 pKa = 9.92SFLLQLLDD227 pKa = 4.18RR228 pKa = 11.84YY229 pKa = 9.19PLRR232 pKa = 11.84VEE234 pKa = 4.21SKK236 pKa = 9.25GGVVQFVGRR245 pKa = 11.84RR246 pKa = 11.84IFITSNAHH254 pKa = 5.41PNQWYY259 pKa = 9.18PNVKK263 pKa = 8.82DD264 pKa = 3.36TAAMFRR270 pKa = 11.84RR271 pKa = 11.84FTRR274 pKa = 11.84IYY276 pKa = 9.8HH277 pKa = 6.05CDD279 pKa = 3.01KK280 pKa = 10.92SRR282 pKa = 11.84FWRR285 pKa = 11.84EE286 pKa = 3.31KK287 pKa = 10.31
Molecular weight: 33.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5U8K1|S5U8K1_9CIRC Capsid protein OS=Farfantepenaeus duorarum circovirus OX=1380894 PE=4 SV=1
MM1 pKa = 7.48NLMLNRR7 pKa = 11.84PRR9 pKa = 11.84LPDD12 pKa = 3.32SSQTKK17 pKa = 9.77GPYY20 pKa = 10.1GYY22 pKa = 10.87AVWANLYY29 pKa = 8.65KK30 pKa = 9.85TVRR33 pKa = 11.84TRR35 pKa = 11.84GVKK38 pKa = 9.69IEE40 pKa = 3.74LWSRR44 pKa = 11.84NRR46 pKa = 11.84SAQPTGSGATFVGFQGSEE64 pKa = 4.01DD65 pKa = 4.94PIPVSDD71 pKa = 3.9TDD73 pKa = 3.88HH74 pKa = 6.63MEE76 pKa = 4.02HH77 pKa = 6.99LLARR81 pKa = 11.84RR82 pKa = 11.84NVIVKK87 pKa = 10.67SNGVPAAGKK96 pKa = 10.15NGSTYY101 pKa = 10.11IKK103 pKa = 10.31AYY105 pKa = 10.42RR106 pKa = 11.84KK107 pKa = 7.84LTNIEE112 pKa = 3.62GHH114 pKa = 5.35QIEE117 pKa = 4.91RR118 pKa = 11.84DD119 pKa = 3.11SDD121 pKa = 3.59YY122 pKa = 11.04WADD125 pKa = 3.85TQQGDD130 pKa = 3.33PAKK133 pKa = 10.18FVKK136 pKa = 10.36AYY138 pKa = 10.72VGVTSAGNVDD148 pKa = 3.54TDD150 pKa = 3.46TSFDD154 pKa = 3.41LTVRR158 pKa = 11.84LTFYY162 pKa = 11.19VDD164 pKa = 3.37FWGIRR169 pKa = 11.84DD170 pKa = 3.77VPSNTLVDD178 pKa = 4.78FAAA181 pKa = 4.78
MM1 pKa = 7.48NLMLNRR7 pKa = 11.84PRR9 pKa = 11.84LPDD12 pKa = 3.32SSQTKK17 pKa = 9.77GPYY20 pKa = 10.1GYY22 pKa = 10.87AVWANLYY29 pKa = 8.65KK30 pKa = 9.85TVRR33 pKa = 11.84TRR35 pKa = 11.84GVKK38 pKa = 9.69IEE40 pKa = 3.74LWSRR44 pKa = 11.84NRR46 pKa = 11.84SAQPTGSGATFVGFQGSEE64 pKa = 4.01DD65 pKa = 4.94PIPVSDD71 pKa = 3.9TDD73 pKa = 3.88HH74 pKa = 6.63MEE76 pKa = 4.02HH77 pKa = 6.99LLARR81 pKa = 11.84RR82 pKa = 11.84NVIVKK87 pKa = 10.67SNGVPAAGKK96 pKa = 10.15NGSTYY101 pKa = 10.11IKK103 pKa = 10.31AYY105 pKa = 10.42RR106 pKa = 11.84KK107 pKa = 7.84LTNIEE112 pKa = 3.62GHH114 pKa = 5.35QIEE117 pKa = 4.91RR118 pKa = 11.84DD119 pKa = 3.11SDD121 pKa = 3.59YY122 pKa = 11.04WADD125 pKa = 3.85TQQGDD130 pKa = 3.33PAKK133 pKa = 10.18FVKK136 pKa = 10.36AYY138 pKa = 10.72VGVTSAGNVDD148 pKa = 3.54TDD150 pKa = 3.46TSFDD154 pKa = 3.41LTVRR158 pKa = 11.84LTFYY162 pKa = 11.19VDD164 pKa = 3.37FWGIRR169 pKa = 11.84DD170 pKa = 3.77VPSNTLVDD178 pKa = 4.78FAAA181 pKa = 4.78
Molecular weight: 20.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
468 |
181 |
287 |
234.0 |
26.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.265 ± 0.275 | 2.137 ± 1.251 |
6.838 ± 0.525 | 4.701 ± 1.134 |
4.06 ± 0.113 | 7.692 ± 0.348 |
2.991 ± 0.781 | 4.06 ± 0.113 |
4.274 ± 0.409 | 6.197 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.923 ± 0.155 | 3.846 ± 0.982 |
3.846 ± 0.659 | 3.632 ± 0.186 |
8.547 ± 1.122 | 6.838 ± 0.202 |
5.342 ± 1.724 | 7.479 ± 0.797 |
3.419 ± 0.707 | 4.915 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
