
Trichoplusia ni (Cabbage looper)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata; Neolepidoptera; Heteroneura; Ditrysia;
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
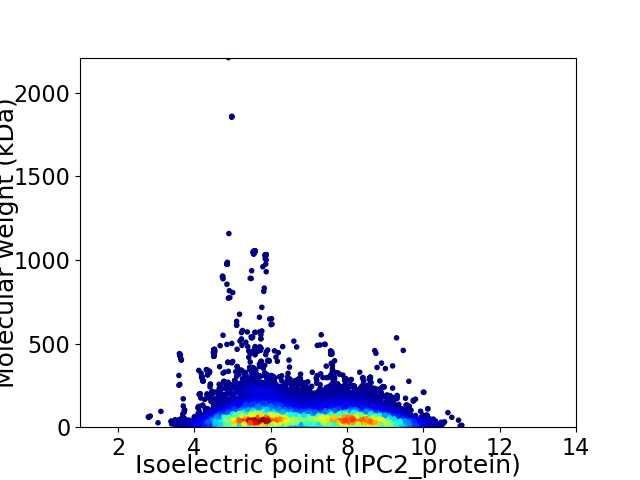
Virtual 2D-PAGE plot for 20889 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7E5WB14|A0A7E5WB14_TRINI pickpocket protein 28 OS=Trichoplusia ni OX=7111 GN=LOC113500897 PE=3 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84SNNTVTDD9 pKa = 3.97SIYY12 pKa = 10.55AYY14 pKa = 10.64FNILSKK20 pKa = 10.77HH21 pKa = 5.59FSQINYY27 pKa = 9.57YY28 pKa = 8.82FICPQIFAYY37 pKa = 9.6FSWWEE42 pKa = 4.02TKK44 pKa = 10.21AIPVPDD50 pKa = 4.75PSASIPSLTTTSTPTTCATDD70 pKa = 3.21STTTSEE76 pKa = 4.19PTTTSDD82 pKa = 4.44SEE84 pKa = 4.61STTEE88 pKa = 5.17AVTTPDD94 pKa = 3.46PQQTTEE100 pKa = 4.04EE101 pKa = 4.52TTTTSPDD108 pKa = 3.18NSPNTTDD115 pKa = 2.87TTTTLDD121 pKa = 3.72PKK123 pKa = 11.41DD124 pKa = 3.65NTTEE128 pKa = 4.07STTTTNEE135 pKa = 4.03TTPEE139 pKa = 4.13TTKK142 pKa = 8.48TTTEE146 pKa = 3.7ATTCNPEE153 pKa = 4.0EE154 pKa = 4.28TTTAATSTTPDD165 pKa = 3.17ANATTEE171 pKa = 4.15PPSTADD177 pKa = 3.1ATATGSTATTPNYY190 pKa = 10.71SDD192 pKa = 3.3TTTEE196 pKa = 4.07TSVNTTDD203 pKa = 3.08STTNKK208 pKa = 10.2PEE210 pKa = 4.17DD211 pKa = 3.62TSTEE215 pKa = 3.75ATATSTEE222 pKa = 4.23SGTTTGATPCSSDD235 pKa = 3.17ATDD238 pKa = 3.45TTTVASATTTGTTPSSSNVTDD259 pKa = 3.59TTLSSATTTEE269 pKa = 3.86SSTDD273 pKa = 3.34SSATPCSSDD282 pKa = 2.88TTTDD286 pKa = 3.17STVTTTSASPCDD298 pKa = 3.98IIEE301 pKa = 4.29TTTDD305 pKa = 2.84SSATTTSTTPCSSDD319 pKa = 3.06AVEE322 pKa = 4.48TTTEE326 pKa = 3.8ASPNITSSPPSATDD340 pKa = 3.18PTEE343 pKa = 3.87PTTDD347 pKa = 3.41SNASTTSATPCSNDD361 pKa = 3.13TTEE364 pKa = 4.29TTPEE368 pKa = 3.95SSLTTTNAAPRR379 pKa = 11.84LTDD382 pKa = 3.43TAEE385 pKa = 4.08TTTDD389 pKa = 2.98SSATTTSATSCSSDD403 pKa = 3.04ASKK406 pKa = 8.09TTTDD410 pKa = 4.13SIATTISAAPCVTDD424 pKa = 3.56TKK426 pKa = 10.64EE427 pKa = 4.03TTTDD431 pKa = 3.14SSATTATATSSEE443 pKa = 4.02YY444 pKa = 10.9DD445 pKa = 3.26ATEE448 pKa = 3.81KK449 pKa = 8.26TTEE452 pKa = 4.13SGVTTTDD459 pKa = 2.97ATPCLSNATQTTTTSTARR477 pKa = 11.84TTDD480 pKa = 3.34ATPCSSDD487 pKa = 3.06ATEE490 pKa = 4.14TTTDD494 pKa = 3.02SSAITTDD501 pKa = 3.26ATPCASDD508 pKa = 3.26ATEE511 pKa = 4.09TTTDD515 pKa = 3.02SSAITTDD522 pKa = 3.12ATPYY526 pKa = 10.97ASDD529 pKa = 3.41ATEE532 pKa = 4.05TTTDD536 pKa = 3.02SSAITTDD543 pKa = 3.26ATPCASDD550 pKa = 3.26ATEE553 pKa = 4.09TTTDD557 pKa = 3.15SSATTTDD564 pKa = 2.96ATPCVSDD571 pKa = 3.46ATEE574 pKa = 4.07TTTDD578 pKa = 3.02SSAITTDD585 pKa = 3.26ATPCASDD592 pKa = 3.26ATEE595 pKa = 4.09TTTDD599 pKa = 3.02SSAITTDD606 pKa = 3.26ATPCASDD613 pKa = 3.32ATEE616 pKa = 3.9TATDD620 pKa = 3.69SSATPTDD627 pKa = 3.62ATPCASDD634 pKa = 3.26ATEE637 pKa = 4.09TTTDD641 pKa = 3.02SSAITTDD648 pKa = 3.26ATPCASDD655 pKa = 3.26ATEE658 pKa = 4.09TTTDD662 pKa = 3.02SSAITTDD669 pKa = 3.26ATPCASDD676 pKa = 3.32ATEE679 pKa = 3.9TATDD683 pKa = 3.67SSATTTDD690 pKa = 3.11ATPCASAATEE700 pKa = 4.31AEE702 pKa = 4.25SDD704 pKa = 3.84STTDD708 pKa = 3.14ATPRR712 pKa = 11.84ASNGDD717 pKa = 3.41TRR719 pKa = 11.84SATSYY724 pKa = 9.92YY725 pKa = 10.37RR726 pKa = 11.84PVGNRR731 pKa = 11.84GNKK734 pKa = 9.23LNHH737 pKa = 6.63ILSKK741 pKa = 10.14PVKK744 pKa = 10.21SLVKK748 pKa = 9.86KK749 pKa = 10.25LRR751 pKa = 11.84PILVFLKK758 pKa = 10.49HH759 pKa = 6.19LSSLLLLFCVQSSPIEE775 pKa = 4.15VKK777 pKa = 9.69MKK779 pKa = 10.49SYY781 pKa = 9.44TLILLVLSCLLVFPSDD797 pKa = 3.68QSALPDD803 pKa = 3.92ASPCFLGPAFYY814 pKa = 9.79PRR816 pKa = 11.84HH817 pKa = 6.1RR818 pKa = 11.84EE819 pKa = 3.65RR820 pKa = 11.84HH821 pKa = 5.47RR822 pKa = 11.84STTTPSTTTSSTSSTSSTTTSTTTSTTSSPQRR854 pKa = 11.84RR855 pKa = 11.84RR856 pKa = 11.84WRR858 pKa = 11.84RR859 pKa = 11.84RR860 pKa = 11.84WFNPFYY866 pKa = 10.05WWWLL870 pKa = 3.45
MM1 pKa = 7.94RR2 pKa = 11.84SNNTVTDD9 pKa = 3.97SIYY12 pKa = 10.55AYY14 pKa = 10.64FNILSKK20 pKa = 10.77HH21 pKa = 5.59FSQINYY27 pKa = 9.57YY28 pKa = 8.82FICPQIFAYY37 pKa = 9.6FSWWEE42 pKa = 4.02TKK44 pKa = 10.21AIPVPDD50 pKa = 4.75PSASIPSLTTTSTPTTCATDD70 pKa = 3.21STTTSEE76 pKa = 4.19PTTTSDD82 pKa = 4.44SEE84 pKa = 4.61STTEE88 pKa = 5.17AVTTPDD94 pKa = 3.46PQQTTEE100 pKa = 4.04EE101 pKa = 4.52TTTTSPDD108 pKa = 3.18NSPNTTDD115 pKa = 2.87TTTTLDD121 pKa = 3.72PKK123 pKa = 11.41DD124 pKa = 3.65NTTEE128 pKa = 4.07STTTTNEE135 pKa = 4.03TTPEE139 pKa = 4.13TTKK142 pKa = 8.48TTTEE146 pKa = 3.7ATTCNPEE153 pKa = 4.0EE154 pKa = 4.28TTTAATSTTPDD165 pKa = 3.17ANATTEE171 pKa = 4.15PPSTADD177 pKa = 3.1ATATGSTATTPNYY190 pKa = 10.71SDD192 pKa = 3.3TTTEE196 pKa = 4.07TSVNTTDD203 pKa = 3.08STTNKK208 pKa = 10.2PEE210 pKa = 4.17DD211 pKa = 3.62TSTEE215 pKa = 3.75ATATSTEE222 pKa = 4.23SGTTTGATPCSSDD235 pKa = 3.17ATDD238 pKa = 3.45TTTVASATTTGTTPSSSNVTDD259 pKa = 3.59TTLSSATTTEE269 pKa = 3.86SSTDD273 pKa = 3.34SSATPCSSDD282 pKa = 2.88TTTDD286 pKa = 3.17STVTTTSASPCDD298 pKa = 3.98IIEE301 pKa = 4.29TTTDD305 pKa = 2.84SSATTTSTTPCSSDD319 pKa = 3.06AVEE322 pKa = 4.48TTTEE326 pKa = 3.8ASPNITSSPPSATDD340 pKa = 3.18PTEE343 pKa = 3.87PTTDD347 pKa = 3.41SNASTTSATPCSNDD361 pKa = 3.13TTEE364 pKa = 4.29TTPEE368 pKa = 3.95SSLTTTNAAPRR379 pKa = 11.84LTDD382 pKa = 3.43TAEE385 pKa = 4.08TTTDD389 pKa = 2.98SSATTTSATSCSSDD403 pKa = 3.04ASKK406 pKa = 8.09TTTDD410 pKa = 4.13SIATTISAAPCVTDD424 pKa = 3.56TKK426 pKa = 10.64EE427 pKa = 4.03TTTDD431 pKa = 3.14SSATTATATSSEE443 pKa = 4.02YY444 pKa = 10.9DD445 pKa = 3.26ATEE448 pKa = 3.81KK449 pKa = 8.26TTEE452 pKa = 4.13SGVTTTDD459 pKa = 2.97ATPCLSNATQTTTTSTARR477 pKa = 11.84TTDD480 pKa = 3.34ATPCSSDD487 pKa = 3.06ATEE490 pKa = 4.14TTTDD494 pKa = 3.02SSAITTDD501 pKa = 3.26ATPCASDD508 pKa = 3.26ATEE511 pKa = 4.09TTTDD515 pKa = 3.02SSAITTDD522 pKa = 3.12ATPYY526 pKa = 10.97ASDD529 pKa = 3.41ATEE532 pKa = 4.05TTTDD536 pKa = 3.02SSAITTDD543 pKa = 3.26ATPCASDD550 pKa = 3.26ATEE553 pKa = 4.09TTTDD557 pKa = 3.15SSATTTDD564 pKa = 2.96ATPCVSDD571 pKa = 3.46ATEE574 pKa = 4.07TTTDD578 pKa = 3.02SSAITTDD585 pKa = 3.26ATPCASDD592 pKa = 3.26ATEE595 pKa = 4.09TTTDD599 pKa = 3.02SSAITTDD606 pKa = 3.26ATPCASDD613 pKa = 3.32ATEE616 pKa = 3.9TATDD620 pKa = 3.69SSATPTDD627 pKa = 3.62ATPCASDD634 pKa = 3.26ATEE637 pKa = 4.09TTTDD641 pKa = 3.02SSAITTDD648 pKa = 3.26ATPCASDD655 pKa = 3.26ATEE658 pKa = 4.09TTTDD662 pKa = 3.02SSAITTDD669 pKa = 3.26ATPCASDD676 pKa = 3.32ATEE679 pKa = 3.9TATDD683 pKa = 3.67SSATTTDD690 pKa = 3.11ATPCASAATEE700 pKa = 4.31AEE702 pKa = 4.25SDD704 pKa = 3.84STTDD708 pKa = 3.14ATPRR712 pKa = 11.84ASNGDD717 pKa = 3.41TRR719 pKa = 11.84SATSYY724 pKa = 9.92YY725 pKa = 10.37RR726 pKa = 11.84PVGNRR731 pKa = 11.84GNKK734 pKa = 9.23LNHH737 pKa = 6.63ILSKK741 pKa = 10.14PVKK744 pKa = 10.21SLVKK748 pKa = 9.86KK749 pKa = 10.25LRR751 pKa = 11.84PILVFLKK758 pKa = 10.49HH759 pKa = 6.19LSSLLLLFCVQSSPIEE775 pKa = 4.15VKK777 pKa = 9.69MKK779 pKa = 10.49SYY781 pKa = 9.44TLILLVLSCLLVFPSDD797 pKa = 3.68QSALPDD803 pKa = 3.92ASPCFLGPAFYY814 pKa = 9.79PRR816 pKa = 11.84HH817 pKa = 6.1RR818 pKa = 11.84EE819 pKa = 3.65RR820 pKa = 11.84HH821 pKa = 5.47RR822 pKa = 11.84STTTPSTTTSSTSSTSSTTTSTTTSTTSSPQRR854 pKa = 11.84RR855 pKa = 11.84RR856 pKa = 11.84WRR858 pKa = 11.84RR859 pKa = 11.84RR860 pKa = 11.84WFNPFYY866 pKa = 10.05WWWLL870 pKa = 3.45
Molecular weight: 89.5 kDa
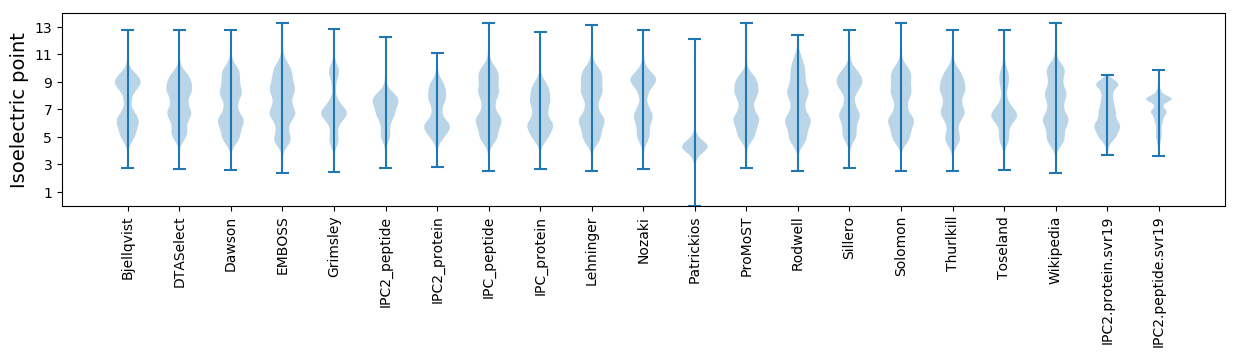
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7E5VNM7|A0A7E5VNM7_TRINI CREB-regulated transcription coactivator 2-like isoform X1 OS=Trichoplusia ni OX=7111 GN=LOC113495410 PE=3 SV=1
MM1 pKa = 7.67ASSKK5 pKa = 9.45TLFILGVLLALVATTFAQNRR25 pKa = 11.84GRR27 pKa = 11.84GGFGGPGGFGGPGGNRR43 pKa = 11.84PGGNRR48 pKa = 11.84PGGGFGGGNNGFGQGGFGQGGFGGGNNGFGPGGFGGGNNGFGQGGFGGNRR98 pKa = 11.84PGGFGGPGGFGGPGGFGRR116 pKa = 11.84PGFGRR121 pKa = 3.64
MM1 pKa = 7.67ASSKK5 pKa = 9.45TLFILGVLLALVATTFAQNRR25 pKa = 11.84GRR27 pKa = 11.84GGFGGPGGFGGPGGNRR43 pKa = 11.84PGGNRR48 pKa = 11.84PGGGFGGGNNGFGQGGFGQGGFGGGNNGFGPGGFGGGNNGFGQGGFGGNRR98 pKa = 11.84PGGFGGPGGFGGPGGFGRR116 pKa = 11.84PGFGRR121 pKa = 3.64
Molecular weight: 11.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13497636 |
34 |
20773 |
646.2 |
72.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.39 ± 0.027 | 1.879 ± 0.025 |
5.628 ± 0.012 | 6.902 ± 0.033 |
3.293 ± 0.014 | 5.655 ± 0.027 |
2.51 ± 0.009 | 4.969 ± 0.019 |
6.353 ± 0.03 | 8.761 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.165 ± 0.01 | 4.53 ± 0.017 |
5.825 ± 0.026 | 4.156 ± 0.021 |
5.733 ± 0.021 | 7.782 ± 0.022 |
6.081 ± 0.041 | 6.324 ± 0.019 |
1.052 ± 0.007 | 3.009 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
