
Tabrizicola piscis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Tabrizicola
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
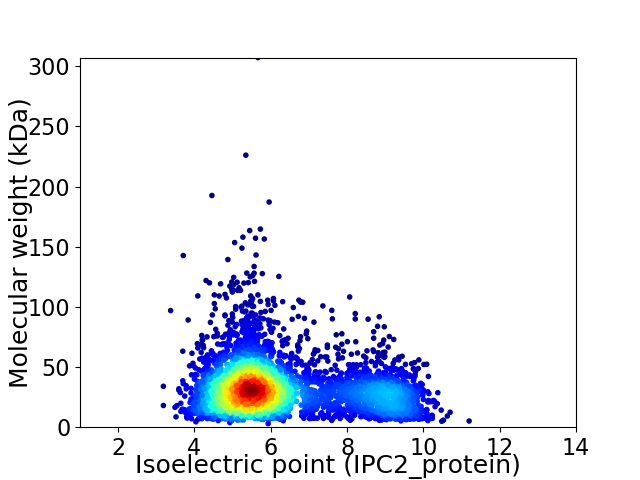
Virtual 2D-PAGE plot for 4119 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UBC0|A0A3S8UBC0_9RHOB Uncharacterized protein OS=Tabrizicola piscis OX=2494374 GN=EI545_20005 PE=4 SV=1
MM1 pKa = 7.57SKK3 pKa = 10.31HH4 pKa = 6.37FSATRR9 pKa = 11.84NNIAVPMPVLEE20 pKa = 4.35TWTLPAALDD29 pKa = 4.23DD30 pKa = 5.22PGLTMADD37 pKa = 4.57FGPVAAPSVTVEE49 pKa = 4.04ATFATSPGCGCVGCSIGTTTEE70 pKa = 4.0TSSSDD75 pKa = 3.09TSGYY79 pKa = 10.87AEE81 pKa = 4.49NGSLATTTSVGLSGNAGIDD100 pKa = 3.39ALLSGIRR107 pKa = 11.84WASTISYY114 pKa = 9.85SDD116 pKa = 3.42PDD118 pKa = 3.46SRR120 pKa = 11.84FDD122 pKa = 3.7YY123 pKa = 10.88QSGYY127 pKa = 10.8YY128 pKa = 9.72VDD130 pKa = 4.8SNGNGSSAQSEE141 pKa = 4.72GFSQISPAMLAALRR155 pKa = 11.84FALDD159 pKa = 3.04ADD161 pKa = 4.25AGVSAARR168 pKa = 11.84GFAIEE173 pKa = 4.45GFTNIKK179 pKa = 9.32IDD181 pKa = 3.73YY182 pKa = 8.98AGSGSGAGTIRR193 pKa = 11.84VANSSDD199 pKa = 3.31APTAYY204 pKa = 10.31AFYY207 pKa = 9.71PDD209 pKa = 3.03GSMYY213 pKa = 10.98GGDD216 pKa = 3.48SFFGTFARR224 pKa = 11.84SPTAAGNYY232 pKa = 8.39SWHH235 pKa = 6.03TMLHH239 pKa = 5.47EE240 pKa = 4.76MGHH243 pKa = 5.57SLGLKK248 pKa = 9.4HH249 pKa = 6.11GHH251 pKa = 5.65EE252 pKa = 4.59FSFGGALPANIDD264 pKa = 3.72SNEE267 pKa = 3.95YY268 pKa = 10.77SVMTYY273 pKa = 10.4RR274 pKa = 11.84SFVGGTTQYY283 pKa = 10.16ATWEE287 pKa = 4.21TWGAPQTYY295 pKa = 9.18MMYY298 pKa = 10.51DD299 pKa = 3.5IAALQHH305 pKa = 6.13MYY307 pKa = 10.77GADD310 pKa = 3.34FSTNSGNTTYY320 pKa = 9.5TWNPTNGQTLINGAVAINPGANRR343 pKa = 11.84IFATIWDD350 pKa = 3.91GGGIDD355 pKa = 4.27TYY357 pKa = 11.67NLSNYY362 pKa = 7.38STNLLIDD369 pKa = 4.04LTPGGHH375 pKa = 5.86SVFSSAQLADD385 pKa = 4.39LNDD388 pKa = 4.14TNWIGLGPQYY398 pKa = 11.49ARR400 pKa = 11.84GNIFNALQFNGDD412 pKa = 3.45SRR414 pKa = 11.84SLIEE418 pKa = 4.06NANGGSGNDD427 pKa = 3.3MLLGNTANNLLNGGAGNDD445 pKa = 3.91TLVGGAGNDD454 pKa = 3.66TYY456 pKa = 11.4VVSSGGDD463 pKa = 3.12QVFEE467 pKa = 4.47TTTTTSTTDD476 pKa = 2.67AGGIDD481 pKa = 4.03TVQSSISFRR490 pKa = 11.84LDD492 pKa = 2.85TTDD495 pKa = 4.28GIRR498 pKa = 11.84FVEE501 pKa = 4.05NLTLTGSGNLSGTGNSLANILTGNAGNNTLFGGLGNDD538 pKa = 4.04TLNGGAGNDD547 pKa = 3.84TLNGGEE553 pKa = 5.33DD554 pKa = 4.25GDD556 pKa = 4.46TMVGGAGNDD565 pKa = 3.65LYY567 pKa = 11.44VVSSTGDD574 pKa = 3.36LVFEE578 pKa = 4.58TTTTTSTTDD587 pKa = 2.67AGGIDD592 pKa = 3.75TVEE595 pKa = 4.24SSVSFNLNANAGVRR609 pKa = 11.84FVEE612 pKa = 3.97NLTLTGTSAVEE623 pKa = 3.85GTGNSRR629 pKa = 11.84ANILTGNSGDD639 pKa = 3.83NILRR643 pKa = 11.84GAAGNDD649 pKa = 3.27TLYY652 pKa = 11.48GLDD655 pKa = 5.2GNDD658 pKa = 3.79TLNGGADD665 pKa = 3.82SDD667 pKa = 4.39TMVGGAGNDD676 pKa = 3.56YY677 pKa = 11.11YY678 pKa = 11.39LVNSTGDD685 pKa = 3.18RR686 pKa = 11.84VFEE689 pKa = 4.42TTTTTSTINAGGIDD703 pKa = 3.99TVQSSVTFSLNTNAGVRR720 pKa = 11.84FVEE723 pKa = 3.88NLTLSGASAVNGTGNSLANTITGNTGANQLNGGLGSDD760 pKa = 3.31ILTGGRR766 pKa = 11.84GADD769 pKa = 2.87SFVFNTALGSGNVDD783 pKa = 4.33RR784 pKa = 11.84ITDD787 pKa = 4.22FDD789 pKa = 3.98VLADD793 pKa = 4.18SIRR796 pKa = 11.84LEE798 pKa = 3.79NAIFIGLSGGTLAASAFAANMTGDD822 pKa = 3.37ATDD825 pKa = 3.13ARR827 pKa = 11.84DD828 pKa = 3.7RR829 pKa = 11.84IIYY832 pKa = 10.23EE833 pKa = 3.9SDD835 pKa = 2.87TGNLYY840 pKa = 10.53FDD842 pKa = 4.0SDD844 pKa = 4.28GTGSAAKK851 pKa = 10.17VHH853 pKa = 6.5FATVDD858 pKa = 3.22TRR860 pKa = 11.84VALTNADD867 pKa = 3.41FFVFF871 pKa = 4.35
MM1 pKa = 7.57SKK3 pKa = 10.31HH4 pKa = 6.37FSATRR9 pKa = 11.84NNIAVPMPVLEE20 pKa = 4.35TWTLPAALDD29 pKa = 4.23DD30 pKa = 5.22PGLTMADD37 pKa = 4.57FGPVAAPSVTVEE49 pKa = 4.04ATFATSPGCGCVGCSIGTTTEE70 pKa = 4.0TSSSDD75 pKa = 3.09TSGYY79 pKa = 10.87AEE81 pKa = 4.49NGSLATTTSVGLSGNAGIDD100 pKa = 3.39ALLSGIRR107 pKa = 11.84WASTISYY114 pKa = 9.85SDD116 pKa = 3.42PDD118 pKa = 3.46SRR120 pKa = 11.84FDD122 pKa = 3.7YY123 pKa = 10.88QSGYY127 pKa = 10.8YY128 pKa = 9.72VDD130 pKa = 4.8SNGNGSSAQSEE141 pKa = 4.72GFSQISPAMLAALRR155 pKa = 11.84FALDD159 pKa = 3.04ADD161 pKa = 4.25AGVSAARR168 pKa = 11.84GFAIEE173 pKa = 4.45GFTNIKK179 pKa = 9.32IDD181 pKa = 3.73YY182 pKa = 8.98AGSGSGAGTIRR193 pKa = 11.84VANSSDD199 pKa = 3.31APTAYY204 pKa = 10.31AFYY207 pKa = 9.71PDD209 pKa = 3.03GSMYY213 pKa = 10.98GGDD216 pKa = 3.48SFFGTFARR224 pKa = 11.84SPTAAGNYY232 pKa = 8.39SWHH235 pKa = 6.03TMLHH239 pKa = 5.47EE240 pKa = 4.76MGHH243 pKa = 5.57SLGLKK248 pKa = 9.4HH249 pKa = 6.11GHH251 pKa = 5.65EE252 pKa = 4.59FSFGGALPANIDD264 pKa = 3.72SNEE267 pKa = 3.95YY268 pKa = 10.77SVMTYY273 pKa = 10.4RR274 pKa = 11.84SFVGGTTQYY283 pKa = 10.16ATWEE287 pKa = 4.21TWGAPQTYY295 pKa = 9.18MMYY298 pKa = 10.51DD299 pKa = 3.5IAALQHH305 pKa = 6.13MYY307 pKa = 10.77GADD310 pKa = 3.34FSTNSGNTTYY320 pKa = 9.5TWNPTNGQTLINGAVAINPGANRR343 pKa = 11.84IFATIWDD350 pKa = 3.91GGGIDD355 pKa = 4.27TYY357 pKa = 11.67NLSNYY362 pKa = 7.38STNLLIDD369 pKa = 4.04LTPGGHH375 pKa = 5.86SVFSSAQLADD385 pKa = 4.39LNDD388 pKa = 4.14TNWIGLGPQYY398 pKa = 11.49ARR400 pKa = 11.84GNIFNALQFNGDD412 pKa = 3.45SRR414 pKa = 11.84SLIEE418 pKa = 4.06NANGGSGNDD427 pKa = 3.3MLLGNTANNLLNGGAGNDD445 pKa = 3.91TLVGGAGNDD454 pKa = 3.66TYY456 pKa = 11.4VVSSGGDD463 pKa = 3.12QVFEE467 pKa = 4.47TTTTTSTTDD476 pKa = 2.67AGGIDD481 pKa = 4.03TVQSSISFRR490 pKa = 11.84LDD492 pKa = 2.85TTDD495 pKa = 4.28GIRR498 pKa = 11.84FVEE501 pKa = 4.05NLTLTGSGNLSGTGNSLANILTGNAGNNTLFGGLGNDD538 pKa = 4.04TLNGGAGNDD547 pKa = 3.84TLNGGEE553 pKa = 5.33DD554 pKa = 4.25GDD556 pKa = 4.46TMVGGAGNDD565 pKa = 3.65LYY567 pKa = 11.44VVSSTGDD574 pKa = 3.36LVFEE578 pKa = 4.58TTTTTSTTDD587 pKa = 2.67AGGIDD592 pKa = 3.75TVEE595 pKa = 4.24SSVSFNLNANAGVRR609 pKa = 11.84FVEE612 pKa = 3.97NLTLTGTSAVEE623 pKa = 3.85GTGNSRR629 pKa = 11.84ANILTGNSGDD639 pKa = 3.83NILRR643 pKa = 11.84GAAGNDD649 pKa = 3.27TLYY652 pKa = 11.48GLDD655 pKa = 5.2GNDD658 pKa = 3.79TLNGGADD665 pKa = 3.82SDD667 pKa = 4.39TMVGGAGNDD676 pKa = 3.56YY677 pKa = 11.11YY678 pKa = 11.39LVNSTGDD685 pKa = 3.18RR686 pKa = 11.84VFEE689 pKa = 4.42TTTTTSTINAGGIDD703 pKa = 3.99TVQSSVTFSLNTNAGVRR720 pKa = 11.84FVEE723 pKa = 3.88NLTLSGASAVNGTGNSLANTITGNTGANQLNGGLGSDD760 pKa = 3.31ILTGGRR766 pKa = 11.84GADD769 pKa = 2.87SFVFNTALGSGNVDD783 pKa = 4.33RR784 pKa = 11.84ITDD787 pKa = 4.22FDD789 pKa = 3.98VLADD793 pKa = 4.18SIRR796 pKa = 11.84LEE798 pKa = 3.79NAIFIGLSGGTLAASAFAANMTGDD822 pKa = 3.37ATDD825 pKa = 3.13ARR827 pKa = 11.84DD828 pKa = 3.7RR829 pKa = 11.84IIYY832 pKa = 10.23EE833 pKa = 3.9SDD835 pKa = 2.87TGNLYY840 pKa = 10.53FDD842 pKa = 4.0SDD844 pKa = 4.28GTGSAAKK851 pKa = 10.17VHH853 pKa = 6.5FATVDD858 pKa = 3.22TRR860 pKa = 11.84VALTNADD867 pKa = 3.41FFVFF871 pKa = 4.35
Molecular weight: 89.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8U5E6|A0A3S8U5E6_9RHOB Serine/threonine protein phosphatase OS=Tabrizicola piscis OX=2494374 GN=EI545_07865 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1303656 |
28 |
2823 |
316.5 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.256 ± 0.049 | 0.839 ± 0.01 |
5.837 ± 0.033 | 5.34 ± 0.031 |
3.628 ± 0.023 | 8.937 ± 0.041 |
2.033 ± 0.019 | 4.865 ± 0.026 |
2.834 ± 0.028 | 10.533 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.02 | 2.28 ± 0.02 |
5.456 ± 0.03 | 3.036 ± 0.019 |
6.871 ± 0.039 | 4.836 ± 0.024 |
5.603 ± 0.026 | 7.561 ± 0.031 |
1.483 ± 0.019 | 2.065 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
