
Nostoc flagelliforme CCNUN1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; Nostoc flagelliforme
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
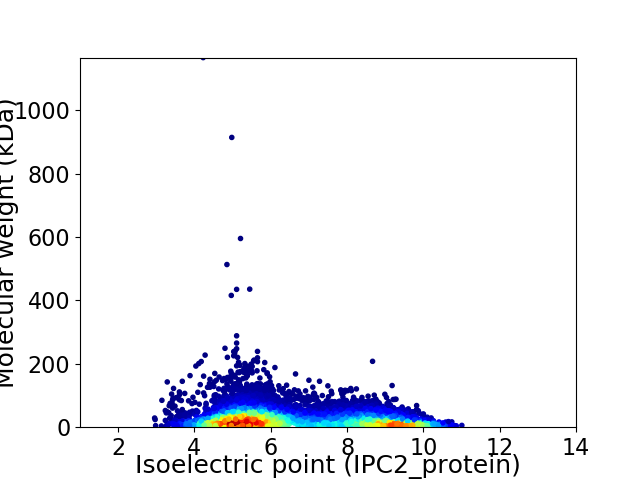
Virtual 2D-PAGE plot for 10244 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
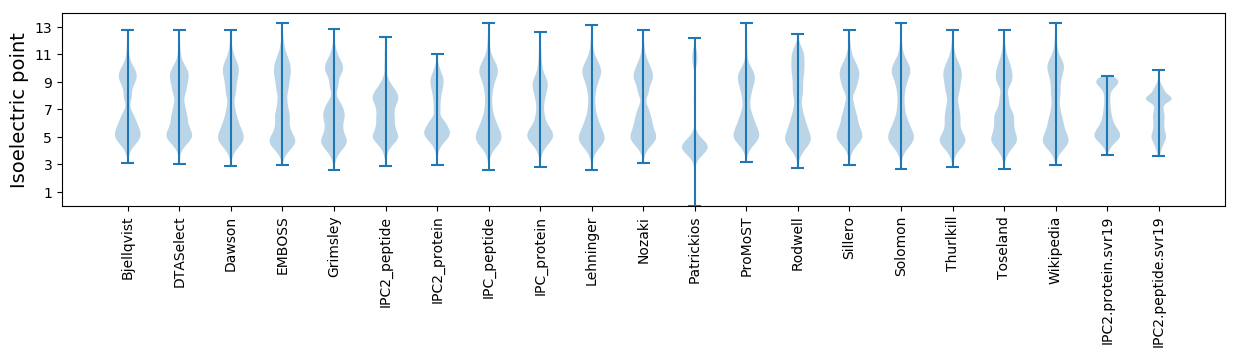
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8T0F6|A0A2K8T0F6_9NOSO Putative transposase YdaD OS=Nostoc flagelliforme CCNUN1 OX=2038116 GN=COO91_07153 PE=4 SV=1
MM1 pKa = 7.32TNIIGSKK8 pKa = 10.92GNDD11 pKa = 3.19TLVGSEE17 pKa = 4.77LADD20 pKa = 4.19TINGLAGNDD29 pKa = 3.94TITATNGNDD38 pKa = 3.19TLTGGGGKK46 pKa = 10.15DD47 pKa = 2.7KK48 pKa = 11.16FKK50 pKa = 11.0FVDD53 pKa = 3.72VFNGTDD59 pKa = 3.39TITDD63 pKa = 4.18FGGVGKK69 pKa = 8.64GTNPTAAVTAEE80 pKa = 3.55VDD82 pKa = 3.42TLKK85 pKa = 11.06FKK87 pKa = 10.9GYY89 pKa = 10.93DD90 pKa = 3.24FFRR93 pKa = 11.84AEE95 pKa = 4.5NLLLTQNGSNLEE107 pKa = 3.93IAFEE111 pKa = 4.51GVGEE115 pKa = 4.31TIVILEE121 pKa = 4.02NFKK124 pKa = 11.28LEE126 pKa = 3.98NLEE129 pKa = 3.99NQKK132 pKa = 11.15ASGATPALGNILFNEE147 pKa = 4.04QTTIIDD153 pKa = 3.64SFDD156 pKa = 3.45VFDD159 pKa = 5.33ANSTEE164 pKa = 4.12TKK166 pKa = 10.51LGIKK170 pKa = 8.81NTVTFLNDD178 pKa = 3.6LSNNITGLDD187 pKa = 3.68DD188 pKa = 4.53SDD190 pKa = 5.48DD191 pKa = 3.87VVNGQGGNDD200 pKa = 3.9KK201 pKa = 10.66IDD203 pKa = 3.71GKK205 pKa = 10.99SGNDD209 pKa = 3.6LLRR212 pKa = 11.84GGTGNDD218 pKa = 3.47TLIGGAGKK226 pKa = 8.59DD227 pKa = 3.5TLIGSTGNNSLLGGAGDD244 pKa = 4.17DD245 pKa = 3.73VLNAEE250 pKa = 4.83DD251 pKa = 3.63STGNSTLNGGAGKK264 pKa = 10.27DD265 pKa = 3.72YY266 pKa = 11.19LSVEE270 pKa = 4.14GSTGDD275 pKa = 3.37NLLFGGDD282 pKa = 3.87GNDD285 pKa = 3.53SFSLRR290 pKa = 11.84ASSTAPSFLVTQTVDD305 pKa = 2.83GGKK308 pKa = 10.65GDD310 pKa = 5.29DD311 pKa = 4.36FLQAYY316 pKa = 6.35YY317 pKa = 10.69TNATGRR323 pKa = 11.84MTSTFNATTNIGSITAGTNRR343 pKa = 11.84VNYY346 pKa = 10.19KK347 pKa = 10.28NIEE350 pKa = 3.97RR351 pKa = 11.84LEE353 pKa = 4.08IRR355 pKa = 11.84GTTYY359 pKa = 11.16NDD361 pKa = 4.72LIVGTNGNDD370 pKa = 3.37YY371 pKa = 11.12LFGGSSGNDD380 pKa = 3.07PSYY383 pKa = 11.9GNDD386 pKa = 3.43TIFGGEE392 pKa = 4.24GNDD395 pKa = 4.24DD396 pKa = 3.95LSVDD400 pKa = 3.92YY401 pKa = 9.85STSDD405 pKa = 3.35NILNGGAGHH414 pKa = 7.08DD415 pKa = 3.73QLSATSSTGNNLLSGDD431 pKa = 4.6DD432 pKa = 4.09GNDD435 pKa = 3.36NLSTSGTHH443 pKa = 6.48IDD445 pKa = 3.67YY446 pKa = 11.23DD447 pKa = 3.93GGNTNAASGNNTLNGGAGDD466 pKa = 4.78DD467 pKa = 4.32NLSADD472 pKa = 3.88YY473 pKa = 9.68STGDD477 pKa = 3.45NLLSGGDD484 pKa = 3.97GNDD487 pKa = 3.39SLSASSYY494 pKa = 9.8PSDD497 pKa = 3.56RR498 pKa = 11.84YY499 pKa = 11.17GFDD502 pKa = 2.97TSGNNTLDD510 pKa = 3.49GGAGDD515 pKa = 4.44DD516 pKa = 3.9TLIANYY522 pKa = 10.52SSGDD526 pKa = 3.44NLFFGGDD533 pKa = 3.64GNDD536 pKa = 3.42SLFVSSYY543 pKa = 9.36YY544 pKa = 10.58DD545 pKa = 3.23YY546 pKa = 11.42SGRR549 pKa = 11.84FDD551 pKa = 3.57GAFGNNLLSGGDD563 pKa = 3.83GNDD566 pKa = 3.24SFSLVLYY573 pKa = 7.18TSPSDD578 pKa = 4.65LVTQTVDD585 pKa = 3.02GGTGDD590 pKa = 4.62DD591 pKa = 4.27LLEE594 pKa = 3.81VSYY597 pKa = 10.52IYY599 pKa = 8.47TTEE602 pKa = 5.14GITSTFDD609 pKa = 3.0AATNTGRR616 pKa = 11.84VTAGTSHH623 pKa = 5.08VTYY626 pKa = 11.04KK627 pKa = 10.86NIEE630 pKa = 3.98GLNISGTEE638 pKa = 3.86YY639 pKa = 11.15DD640 pKa = 4.1DD641 pKa = 6.65LIVGSNGNDD650 pKa = 3.52KK651 pKa = 10.66IDD653 pKa = 3.76GSVSGKK659 pKa = 8.76DD660 pKa = 3.47TIDD663 pKa = 3.5GGTGDD668 pKa = 5.03DD669 pKa = 4.02YY670 pKa = 11.94LSFSCYY676 pKa = 10.2SATQSITSTFDD687 pKa = 2.84AATNTGRR694 pKa = 11.84VTAGTSHH701 pKa = 5.08VTYY704 pKa = 11.09KK705 pKa = 10.86NIEE708 pKa = 3.83QLNIYY713 pKa = 8.28GTDD716 pKa = 3.58YY717 pKa = 11.68DD718 pKa = 4.26DD719 pKa = 7.04LIVGSNGNDD728 pKa = 3.31SLHH731 pKa = 6.65GGNSGNDD738 pKa = 3.57TIDD741 pKa = 3.37GGKK744 pKa = 10.57GDD746 pKa = 5.43DD747 pKa = 4.99LLSLYY752 pKa = 10.76NYY754 pKa = 6.93TTSDD758 pKa = 3.97GITSTFNAATNTGRR772 pKa = 11.84VTAGTSHH779 pKa = 5.08VTYY782 pKa = 11.09KK783 pKa = 10.86NIEE786 pKa = 3.88QLNISGTEE794 pKa = 3.8YY795 pKa = 11.15DD796 pKa = 4.1DD797 pKa = 6.65LIVGSNGNDD806 pKa = 3.79TINGAGGNNTIFGGAGKK823 pKa = 10.17DD824 pKa = 3.75YY825 pKa = 11.34LSADD829 pKa = 3.51YY830 pKa = 11.67SNGNNLLSGDD840 pKa = 4.74DD841 pKa = 4.67GNDD844 pKa = 3.21SLNISGHH851 pKa = 5.17YY852 pKa = 9.63NDD854 pKa = 4.67YY855 pKa = 10.85RR856 pKa = 11.84GEE858 pKa = 4.17FYY860 pKa = 11.18LDD862 pKa = 3.05AVSGNNTLNGGAGDD876 pKa = 4.58DD877 pKa = 3.79NLVTNYY883 pKa = 10.67SSGNNLLSGGDD894 pKa = 3.63GNDD897 pKa = 3.42YY898 pKa = 11.12LSASGSASLYY908 pKa = 10.15SSYY911 pKa = 11.34GVYY914 pKa = 9.2TVSGNNTLNGGAGNDD929 pKa = 3.6RR930 pKa = 11.84LIVDD934 pKa = 4.29YY935 pKa = 10.22STGDD939 pKa = 3.41NLLSGGDD946 pKa = 3.92GNDD949 pKa = 3.03SLTAFGASGKK959 pKa = 8.37NTLTGGNGNDD969 pKa = 3.58TLTGGKK975 pKa = 10.55GNDD978 pKa = 3.62SLIGGAGTDD987 pKa = 3.52TFALNSFNEE996 pKa = 5.19GIDD999 pKa = 3.49TIYY1002 pKa = 11.11DD1003 pKa = 3.76FNATNEE1009 pKa = 4.49LIQISAAGFGGGLSSGSLEE1028 pKa = 4.13TSHH1031 pKa = 6.51FTLGTSATTSEE1042 pKa = 3.85EE1043 pKa = 3.77RR1044 pKa = 11.84FIYY1047 pKa = 10.64NSATGGLFFDD1057 pKa = 5.26LDD1059 pKa = 3.95GSASGSTQVQFAQLSAGLSVTEE1081 pKa = 4.07NNFVVVV1087 pKa = 4.06
MM1 pKa = 7.32TNIIGSKK8 pKa = 10.92GNDD11 pKa = 3.19TLVGSEE17 pKa = 4.77LADD20 pKa = 4.19TINGLAGNDD29 pKa = 3.94TITATNGNDD38 pKa = 3.19TLTGGGGKK46 pKa = 10.15DD47 pKa = 2.7KK48 pKa = 11.16FKK50 pKa = 11.0FVDD53 pKa = 3.72VFNGTDD59 pKa = 3.39TITDD63 pKa = 4.18FGGVGKK69 pKa = 8.64GTNPTAAVTAEE80 pKa = 3.55VDD82 pKa = 3.42TLKK85 pKa = 11.06FKK87 pKa = 10.9GYY89 pKa = 10.93DD90 pKa = 3.24FFRR93 pKa = 11.84AEE95 pKa = 4.5NLLLTQNGSNLEE107 pKa = 3.93IAFEE111 pKa = 4.51GVGEE115 pKa = 4.31TIVILEE121 pKa = 4.02NFKK124 pKa = 11.28LEE126 pKa = 3.98NLEE129 pKa = 3.99NQKK132 pKa = 11.15ASGATPALGNILFNEE147 pKa = 4.04QTTIIDD153 pKa = 3.64SFDD156 pKa = 3.45VFDD159 pKa = 5.33ANSTEE164 pKa = 4.12TKK166 pKa = 10.51LGIKK170 pKa = 8.81NTVTFLNDD178 pKa = 3.6LSNNITGLDD187 pKa = 3.68DD188 pKa = 4.53SDD190 pKa = 5.48DD191 pKa = 3.87VVNGQGGNDD200 pKa = 3.9KK201 pKa = 10.66IDD203 pKa = 3.71GKK205 pKa = 10.99SGNDD209 pKa = 3.6LLRR212 pKa = 11.84GGTGNDD218 pKa = 3.47TLIGGAGKK226 pKa = 8.59DD227 pKa = 3.5TLIGSTGNNSLLGGAGDD244 pKa = 4.17DD245 pKa = 3.73VLNAEE250 pKa = 4.83DD251 pKa = 3.63STGNSTLNGGAGKK264 pKa = 10.27DD265 pKa = 3.72YY266 pKa = 11.19LSVEE270 pKa = 4.14GSTGDD275 pKa = 3.37NLLFGGDD282 pKa = 3.87GNDD285 pKa = 3.53SFSLRR290 pKa = 11.84ASSTAPSFLVTQTVDD305 pKa = 2.83GGKK308 pKa = 10.65GDD310 pKa = 5.29DD311 pKa = 4.36FLQAYY316 pKa = 6.35YY317 pKa = 10.69TNATGRR323 pKa = 11.84MTSTFNATTNIGSITAGTNRR343 pKa = 11.84VNYY346 pKa = 10.19KK347 pKa = 10.28NIEE350 pKa = 3.97RR351 pKa = 11.84LEE353 pKa = 4.08IRR355 pKa = 11.84GTTYY359 pKa = 11.16NDD361 pKa = 4.72LIVGTNGNDD370 pKa = 3.37YY371 pKa = 11.12LFGGSSGNDD380 pKa = 3.07PSYY383 pKa = 11.9GNDD386 pKa = 3.43TIFGGEE392 pKa = 4.24GNDD395 pKa = 4.24DD396 pKa = 3.95LSVDD400 pKa = 3.92YY401 pKa = 9.85STSDD405 pKa = 3.35NILNGGAGHH414 pKa = 7.08DD415 pKa = 3.73QLSATSSTGNNLLSGDD431 pKa = 4.6DD432 pKa = 4.09GNDD435 pKa = 3.36NLSTSGTHH443 pKa = 6.48IDD445 pKa = 3.67YY446 pKa = 11.23DD447 pKa = 3.93GGNTNAASGNNTLNGGAGDD466 pKa = 4.78DD467 pKa = 4.32NLSADD472 pKa = 3.88YY473 pKa = 9.68STGDD477 pKa = 3.45NLLSGGDD484 pKa = 3.97GNDD487 pKa = 3.39SLSASSYY494 pKa = 9.8PSDD497 pKa = 3.56RR498 pKa = 11.84YY499 pKa = 11.17GFDD502 pKa = 2.97TSGNNTLDD510 pKa = 3.49GGAGDD515 pKa = 4.44DD516 pKa = 3.9TLIANYY522 pKa = 10.52SSGDD526 pKa = 3.44NLFFGGDD533 pKa = 3.64GNDD536 pKa = 3.42SLFVSSYY543 pKa = 9.36YY544 pKa = 10.58DD545 pKa = 3.23YY546 pKa = 11.42SGRR549 pKa = 11.84FDD551 pKa = 3.57GAFGNNLLSGGDD563 pKa = 3.83GNDD566 pKa = 3.24SFSLVLYY573 pKa = 7.18TSPSDD578 pKa = 4.65LVTQTVDD585 pKa = 3.02GGTGDD590 pKa = 4.62DD591 pKa = 4.27LLEE594 pKa = 3.81VSYY597 pKa = 10.52IYY599 pKa = 8.47TTEE602 pKa = 5.14GITSTFDD609 pKa = 3.0AATNTGRR616 pKa = 11.84VTAGTSHH623 pKa = 5.08VTYY626 pKa = 11.04KK627 pKa = 10.86NIEE630 pKa = 3.98GLNISGTEE638 pKa = 3.86YY639 pKa = 11.15DD640 pKa = 4.1DD641 pKa = 6.65LIVGSNGNDD650 pKa = 3.52KK651 pKa = 10.66IDD653 pKa = 3.76GSVSGKK659 pKa = 8.76DD660 pKa = 3.47TIDD663 pKa = 3.5GGTGDD668 pKa = 5.03DD669 pKa = 4.02YY670 pKa = 11.94LSFSCYY676 pKa = 10.2SATQSITSTFDD687 pKa = 2.84AATNTGRR694 pKa = 11.84VTAGTSHH701 pKa = 5.08VTYY704 pKa = 11.09KK705 pKa = 10.86NIEE708 pKa = 3.83QLNIYY713 pKa = 8.28GTDD716 pKa = 3.58YY717 pKa = 11.68DD718 pKa = 4.26DD719 pKa = 7.04LIVGSNGNDD728 pKa = 3.31SLHH731 pKa = 6.65GGNSGNDD738 pKa = 3.57TIDD741 pKa = 3.37GGKK744 pKa = 10.57GDD746 pKa = 5.43DD747 pKa = 4.99LLSLYY752 pKa = 10.76NYY754 pKa = 6.93TTSDD758 pKa = 3.97GITSTFNAATNTGRR772 pKa = 11.84VTAGTSHH779 pKa = 5.08VTYY782 pKa = 11.09KK783 pKa = 10.86NIEE786 pKa = 3.88QLNISGTEE794 pKa = 3.8YY795 pKa = 11.15DD796 pKa = 4.1DD797 pKa = 6.65LIVGSNGNDD806 pKa = 3.79TINGAGGNNTIFGGAGKK823 pKa = 10.17DD824 pKa = 3.75YY825 pKa = 11.34LSADD829 pKa = 3.51YY830 pKa = 11.67SNGNNLLSGDD840 pKa = 4.74DD841 pKa = 4.67GNDD844 pKa = 3.21SLNISGHH851 pKa = 5.17YY852 pKa = 9.63NDD854 pKa = 4.67YY855 pKa = 10.85RR856 pKa = 11.84GEE858 pKa = 4.17FYY860 pKa = 11.18LDD862 pKa = 3.05AVSGNNTLNGGAGDD876 pKa = 4.58DD877 pKa = 3.79NLVTNYY883 pKa = 10.67SSGNNLLSGGDD894 pKa = 3.63GNDD897 pKa = 3.42YY898 pKa = 11.12LSASGSASLYY908 pKa = 10.15SSYY911 pKa = 11.34GVYY914 pKa = 9.2TVSGNNTLNGGAGNDD929 pKa = 3.6RR930 pKa = 11.84LIVDD934 pKa = 4.29YY935 pKa = 10.22STGDD939 pKa = 3.41NLLSGGDD946 pKa = 3.92GNDD949 pKa = 3.03SLTAFGASGKK959 pKa = 8.37NTLTGGNGNDD969 pKa = 3.58TLTGGKK975 pKa = 10.55GNDD978 pKa = 3.62SLIGGAGTDD987 pKa = 3.52TFALNSFNEE996 pKa = 5.19GIDD999 pKa = 3.49TIYY1002 pKa = 11.11DD1003 pKa = 3.76FNATNEE1009 pKa = 4.49LIQISAAGFGGGLSSGSLEE1028 pKa = 4.13TSHH1031 pKa = 6.51FTLGTSATTSEE1042 pKa = 3.85EE1043 pKa = 3.77RR1044 pKa = 11.84FIYY1047 pKa = 10.64NSATGGLFFDD1057 pKa = 5.26LDD1059 pKa = 3.95GSASGSTQVQFAQLSAGLSVTEE1081 pKa = 4.07NNFVVVV1087 pKa = 4.06
Molecular weight: 111.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8SHK8|A0A2K8SHK8_9NOSO Glucan-binding domain OS=Nostoc flagelliforme CCNUN1 OX=2038116 GN=COO91_00759 PE=4 SV=1
MM1 pKa = 7.44SLFARR6 pKa = 11.84QLNLFARR13 pKa = 11.84QLSLFARR20 pKa = 11.84QLSLFARR27 pKa = 11.84QLSLFARR34 pKa = 11.84QLSLFTRR41 pKa = 11.84QLSLFTRR48 pKa = 4.37
MM1 pKa = 7.44SLFARR6 pKa = 11.84QLNLFARR13 pKa = 11.84QLSLFARR20 pKa = 11.84QLSLFARR27 pKa = 11.84QLSLFARR34 pKa = 11.84QLSLFTRR41 pKa = 11.84QLSLFTRR48 pKa = 4.37
Molecular weight: 5.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2470546 |
29 |
10889 |
241.2 |
26.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.039 ± 0.031 | 1.008 ± 0.01 |
4.839 ± 0.023 | 6.274 ± 0.031 |
3.964 ± 0.019 | 6.629 ± 0.037 |
1.784 ± 0.013 | 6.724 ± 0.023 |
5.076 ± 0.03 | 10.878 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.815 ± 0.014 | 4.602 ± 0.033 |
4.594 ± 0.021 | 5.323 ± 0.028 |
5.096 ± 0.023 | 6.712 ± 0.025 |
5.717 ± 0.028 | 6.504 ± 0.022 |
1.408 ± 0.012 | 3.014 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
