
Trichoplax adhaerens (Trichoplax reptans)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Placozoa; Trichoplacidae; Trichoplax
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
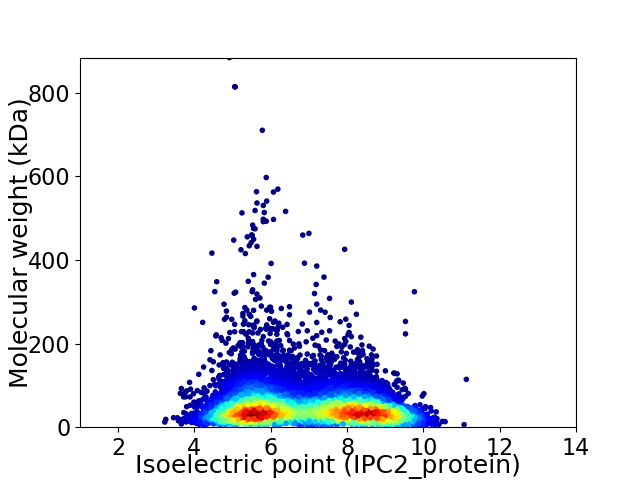
Virtual 2D-PAGE plot for 11520 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3RQ32|B3RQ32_TRIAD 2-Hacid_dh_C domain-containing protein OS=Trichoplax adhaerens OX=10228 GN=TRIADDRAFT_53759 PE=4 SV=1
MM1 pKa = 6.47YY2 pKa = 9.56TPEE5 pKa = 5.08ASIQTEE11 pKa = 3.82SDD13 pKa = 3.34IIADD17 pKa = 3.24ITSQDD22 pKa = 3.44EE23 pKa = 4.19NLKK26 pKa = 8.21EE27 pKa = 4.2TRR29 pKa = 11.84STILLSDD36 pKa = 3.4VDD38 pKa = 4.3IEE40 pKa = 4.25LSSTIGNEE48 pKa = 4.14EE49 pKa = 3.89EE50 pKa = 5.06DD51 pKa = 4.27KK52 pKa = 11.27EE53 pKa = 4.5STNIDD58 pKa = 3.83NIISATDD65 pKa = 3.25IQEE68 pKa = 4.01SSKK71 pKa = 10.99DD72 pKa = 3.4QSILHH77 pKa = 6.12TLQSTIDD84 pKa = 3.68NFEE87 pKa = 3.88VSSPVNNIEE96 pKa = 4.15SSSIDD101 pKa = 3.4IISDD105 pKa = 3.36KK106 pKa = 11.06VSIYY110 pKa = 10.87SDD112 pKa = 3.57GLLQTITQNVMYY124 pKa = 9.19TPEE127 pKa = 4.68ASIQTEE133 pKa = 3.82SDD135 pKa = 3.34IIADD139 pKa = 3.24ITSQDD144 pKa = 3.44EE145 pKa = 4.19NLKK148 pKa = 8.21EE149 pKa = 4.19TRR151 pKa = 11.84STILISDD158 pKa = 3.52VDD160 pKa = 3.88IEE162 pKa = 4.21LSSTIGNEE170 pKa = 4.01EE171 pKa = 4.11EE172 pKa = 4.48EE173 pKa = 4.5KK174 pKa = 10.91EE175 pKa = 4.22STNIDD180 pKa = 3.84NIISATDD187 pKa = 3.25IQEE190 pKa = 4.01SSKK193 pKa = 10.98DD194 pKa = 3.47QSILYY199 pKa = 7.97TLQSTIDD206 pKa = 3.65NFEE209 pKa = 3.88VSSPVNNIEE218 pKa = 4.37SNSIDD223 pKa = 3.69IISDD227 pKa = 3.69KK228 pKa = 10.87ISIYY232 pKa = 10.8SDD234 pKa = 3.16GLSQTNTQNIMYY246 pKa = 8.7TPEE249 pKa = 4.55ASIQTEE255 pKa = 3.82SDD257 pKa = 3.34IIADD261 pKa = 3.23ITSQDD266 pKa = 2.85EE267 pKa = 4.33HH268 pKa = 8.04LQEE271 pKa = 4.45TQSTTLISDD280 pKa = 3.51VSIEE284 pKa = 4.09LSNTIQSEE292 pKa = 4.55EE293 pKa = 4.11EE294 pKa = 3.8KK295 pKa = 9.96STNIDD300 pKa = 3.25NSILTTDD307 pKa = 3.27IQEE310 pKa = 4.28SSEE313 pKa = 4.07DD314 pKa = 3.58QPILHH319 pKa = 6.77TLQSTIDD326 pKa = 3.53NFKK329 pKa = 10.45ISSSVDD335 pKa = 3.36NIEE338 pKa = 4.49SNSIDD343 pKa = 3.59IISDD347 pKa = 3.33KK348 pKa = 11.06VSIYY352 pKa = 10.87SDD354 pKa = 3.57GLLQTITQNIMYY366 pKa = 8.73TPEE369 pKa = 4.55ASIQTGSDD377 pKa = 3.36IIADD381 pKa = 3.18ITSQDD386 pKa = 3.27EE387 pKa = 4.13NLQEE391 pKa = 4.27TRR393 pKa = 11.84STTLISDD400 pKa = 3.43VDD402 pKa = 3.73IEE404 pKa = 4.21LSSTIGNEE412 pKa = 4.01EE413 pKa = 4.11EE414 pKa = 4.48EE415 pKa = 4.5KK416 pKa = 10.91EE417 pKa = 4.22STNIDD422 pKa = 3.78NIISPTGGNEE432 pKa = 3.6IVVTKK437 pKa = 10.65SIGFMYY443 pKa = 10.4HH444 pKa = 6.86SSDD447 pKa = 3.21QSAVTDD453 pKa = 3.53EE454 pKa = 4.42FSSVEE459 pKa = 3.84IFNNALSISVNHH471 pKa = 6.65SNFQTIQEE479 pKa = 4.14NSDD482 pKa = 3.77FEE484 pKa = 4.4LSNIISTNGYY494 pKa = 10.31ASLPPIEE501 pKa = 5.85LSTQEE506 pKa = 3.8YY507 pKa = 10.45ASRR510 pKa = 11.84SNHH513 pKa = 4.61EE514 pKa = 4.3TYY516 pKa = 11.08LLQASSDD523 pKa = 4.21SINGFTMFNSLSSLVDD539 pKa = 3.65SVSSNGNFEE548 pKa = 4.56QIASVPEE555 pKa = 4.0SGISSMEE562 pKa = 4.11SVNDD566 pKa = 3.28AVTFTNAVGPFQSIFNSRR584 pKa = 11.84NDD586 pKa = 3.0ISLSQWNNIAGASYY600 pKa = 11.13AVTTSDD606 pKa = 3.76PLIAFVGSVSSDD618 pKa = 3.27HH619 pKa = 6.35YY620 pKa = 11.1SHH622 pKa = 7.59TINHH626 pKa = 5.53EE627 pKa = 4.24TQSILIMASTDD638 pKa = 3.16SSSEE642 pKa = 4.39SNSSAPDD649 pKa = 4.23DD650 pKa = 3.78IYY652 pKa = 11.62SEE654 pKa = 4.07IRR656 pKa = 11.84GSISSSAVDD665 pKa = 3.31TLTISMNNYY674 pKa = 10.41SMLSSSTYY682 pKa = 9.98PFPLDD687 pKa = 3.28PHH689 pKa = 5.92EE690 pKa = 4.73TQSSTHH696 pKa = 3.87THH698 pKa = 5.47YY699 pKa = 11.28GEE701 pKa = 4.51SYY703 pKa = 9.66RR704 pKa = 11.84EE705 pKa = 4.11SPASILLDD713 pKa = 3.24HH714 pKa = 7.09KK715 pKa = 11.6SNVTSFVQSSNLAQIVNVTGSDD737 pKa = 3.29GLDD740 pKa = 3.21NVEE743 pKa = 4.15TSSSIVAFSPTFFSSFYY760 pKa = 10.86SGDD763 pKa = 3.27GNIYY767 pKa = 10.13DD768 pKa = 3.73QSSTNSMIDD777 pKa = 3.52DD778 pKa = 4.91NIAPEE783 pKa = 4.16PSSVLEE789 pKa = 3.95NTEE792 pKa = 4.48TISKK796 pKa = 8.89QQTIEE801 pKa = 4.12DD802 pKa = 4.42CSTNMNLCDD811 pKa = 3.72TNAICTNTLGSFICSCVTGFTGNGTFCEE839 pKa = 4.45GKK841 pKa = 10.04EE842 pKa = 3.95FQSLYY847 pKa = 11.0LHH849 pKa = 6.82FLL851 pKa = 3.66
MM1 pKa = 6.47YY2 pKa = 9.56TPEE5 pKa = 5.08ASIQTEE11 pKa = 3.82SDD13 pKa = 3.34IIADD17 pKa = 3.24ITSQDD22 pKa = 3.44EE23 pKa = 4.19NLKK26 pKa = 8.21EE27 pKa = 4.2TRR29 pKa = 11.84STILLSDD36 pKa = 3.4VDD38 pKa = 4.3IEE40 pKa = 4.25LSSTIGNEE48 pKa = 4.14EE49 pKa = 3.89EE50 pKa = 5.06DD51 pKa = 4.27KK52 pKa = 11.27EE53 pKa = 4.5STNIDD58 pKa = 3.83NIISATDD65 pKa = 3.25IQEE68 pKa = 4.01SSKK71 pKa = 10.99DD72 pKa = 3.4QSILHH77 pKa = 6.12TLQSTIDD84 pKa = 3.68NFEE87 pKa = 3.88VSSPVNNIEE96 pKa = 4.15SSSIDD101 pKa = 3.4IISDD105 pKa = 3.36KK106 pKa = 11.06VSIYY110 pKa = 10.87SDD112 pKa = 3.57GLLQTITQNVMYY124 pKa = 9.19TPEE127 pKa = 4.68ASIQTEE133 pKa = 3.82SDD135 pKa = 3.34IIADD139 pKa = 3.24ITSQDD144 pKa = 3.44EE145 pKa = 4.19NLKK148 pKa = 8.21EE149 pKa = 4.19TRR151 pKa = 11.84STILISDD158 pKa = 3.52VDD160 pKa = 3.88IEE162 pKa = 4.21LSSTIGNEE170 pKa = 4.01EE171 pKa = 4.11EE172 pKa = 4.48EE173 pKa = 4.5KK174 pKa = 10.91EE175 pKa = 4.22STNIDD180 pKa = 3.84NIISATDD187 pKa = 3.25IQEE190 pKa = 4.01SSKK193 pKa = 10.98DD194 pKa = 3.47QSILYY199 pKa = 7.97TLQSTIDD206 pKa = 3.65NFEE209 pKa = 3.88VSSPVNNIEE218 pKa = 4.37SNSIDD223 pKa = 3.69IISDD227 pKa = 3.69KK228 pKa = 10.87ISIYY232 pKa = 10.8SDD234 pKa = 3.16GLSQTNTQNIMYY246 pKa = 8.7TPEE249 pKa = 4.55ASIQTEE255 pKa = 3.82SDD257 pKa = 3.34IIADD261 pKa = 3.23ITSQDD266 pKa = 2.85EE267 pKa = 4.33HH268 pKa = 8.04LQEE271 pKa = 4.45TQSTTLISDD280 pKa = 3.51VSIEE284 pKa = 4.09LSNTIQSEE292 pKa = 4.55EE293 pKa = 4.11EE294 pKa = 3.8KK295 pKa = 9.96STNIDD300 pKa = 3.25NSILTTDD307 pKa = 3.27IQEE310 pKa = 4.28SSEE313 pKa = 4.07DD314 pKa = 3.58QPILHH319 pKa = 6.77TLQSTIDD326 pKa = 3.53NFKK329 pKa = 10.45ISSSVDD335 pKa = 3.36NIEE338 pKa = 4.49SNSIDD343 pKa = 3.59IISDD347 pKa = 3.33KK348 pKa = 11.06VSIYY352 pKa = 10.87SDD354 pKa = 3.57GLLQTITQNIMYY366 pKa = 8.73TPEE369 pKa = 4.55ASIQTGSDD377 pKa = 3.36IIADD381 pKa = 3.18ITSQDD386 pKa = 3.27EE387 pKa = 4.13NLQEE391 pKa = 4.27TRR393 pKa = 11.84STTLISDD400 pKa = 3.43VDD402 pKa = 3.73IEE404 pKa = 4.21LSSTIGNEE412 pKa = 4.01EE413 pKa = 4.11EE414 pKa = 4.48EE415 pKa = 4.5KK416 pKa = 10.91EE417 pKa = 4.22STNIDD422 pKa = 3.78NIISPTGGNEE432 pKa = 3.6IVVTKK437 pKa = 10.65SIGFMYY443 pKa = 10.4HH444 pKa = 6.86SSDD447 pKa = 3.21QSAVTDD453 pKa = 3.53EE454 pKa = 4.42FSSVEE459 pKa = 3.84IFNNALSISVNHH471 pKa = 6.65SNFQTIQEE479 pKa = 4.14NSDD482 pKa = 3.77FEE484 pKa = 4.4LSNIISTNGYY494 pKa = 10.31ASLPPIEE501 pKa = 5.85LSTQEE506 pKa = 3.8YY507 pKa = 10.45ASRR510 pKa = 11.84SNHH513 pKa = 4.61EE514 pKa = 4.3TYY516 pKa = 11.08LLQASSDD523 pKa = 4.21SINGFTMFNSLSSLVDD539 pKa = 3.65SVSSNGNFEE548 pKa = 4.56QIASVPEE555 pKa = 4.0SGISSMEE562 pKa = 4.11SVNDD566 pKa = 3.28AVTFTNAVGPFQSIFNSRR584 pKa = 11.84NDD586 pKa = 3.0ISLSQWNNIAGASYY600 pKa = 11.13AVTTSDD606 pKa = 3.76PLIAFVGSVSSDD618 pKa = 3.27HH619 pKa = 6.35YY620 pKa = 11.1SHH622 pKa = 7.59TINHH626 pKa = 5.53EE627 pKa = 4.24TQSILIMASTDD638 pKa = 3.16SSSEE642 pKa = 4.39SNSSAPDD649 pKa = 4.23DD650 pKa = 3.78IYY652 pKa = 11.62SEE654 pKa = 4.07IRR656 pKa = 11.84GSISSSAVDD665 pKa = 3.31TLTISMNNYY674 pKa = 10.41SMLSSSTYY682 pKa = 9.98PFPLDD687 pKa = 3.28PHH689 pKa = 5.92EE690 pKa = 4.73TQSSTHH696 pKa = 3.87THH698 pKa = 5.47YY699 pKa = 11.28GEE701 pKa = 4.51SYY703 pKa = 9.66RR704 pKa = 11.84EE705 pKa = 4.11SPASILLDD713 pKa = 3.24HH714 pKa = 7.09KK715 pKa = 11.6SNVTSFVQSSNLAQIVNVTGSDD737 pKa = 3.29GLDD740 pKa = 3.21NVEE743 pKa = 4.15TSSSIVAFSPTFFSSFYY760 pKa = 10.86SGDD763 pKa = 3.27GNIYY767 pKa = 10.13DD768 pKa = 3.73QSSTNSMIDD777 pKa = 3.52DD778 pKa = 4.91NIAPEE783 pKa = 4.16PSSVLEE789 pKa = 3.95NTEE792 pKa = 4.48TISKK796 pKa = 8.89QQTIEE801 pKa = 4.12DD802 pKa = 4.42CSTNMNLCDD811 pKa = 3.72TNAICTNTLGSFICSCVTGFTGNGTFCEE839 pKa = 4.45GKK841 pKa = 10.04EE842 pKa = 3.95FQSLYY847 pKa = 11.0LHH849 pKa = 6.82FLL851 pKa = 3.66
Molecular weight: 92.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3RZS5|B3RZS5_TRIAD Glyco_tran_10_N domain-containing protein OS=Trichoplax adhaerens OX=10228 GN=TRIADDRAFT_26273 PE=3 SV=1
SS1 pKa = 6.89HH2 pKa = 7.64KK3 pKa = 11.01SFIIKK8 pKa = 10.17RR9 pKa = 11.84KK10 pKa = 8.59LAKK13 pKa = 10.03KK14 pKa = 8.72MKK16 pKa = 9.55QNRR19 pKa = 11.84PVPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TNNKK32 pKa = 8.55IRR34 pKa = 11.84YY35 pKa = 5.25NTKK38 pKa = 8.17RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 3.92WRR43 pKa = 11.84RR44 pKa = 11.84TKK46 pKa = 10.83LGLL49 pKa = 3.67
SS1 pKa = 6.89HH2 pKa = 7.64KK3 pKa = 11.01SFIIKK8 pKa = 10.17RR9 pKa = 11.84KK10 pKa = 8.59LAKK13 pKa = 10.03KK14 pKa = 8.72MKK16 pKa = 9.55QNRR19 pKa = 11.84PVPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TNNKK32 pKa = 8.55IRR34 pKa = 11.84YY35 pKa = 5.25NTKK38 pKa = 8.17RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 3.92WRR43 pKa = 11.84RR44 pKa = 11.84TKK46 pKa = 10.83LGLL49 pKa = 3.67
Molecular weight: 6.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5222991 |
49 |
7710 |
453.4 |
51.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.994 ± 0.021 | 2.096 ± 0.027 |
5.688 ± 0.016 | 5.807 ± 0.029 |
4.103 ± 0.017 | 5.071 ± 0.028 |
2.342 ± 0.011 | 7.327 ± 0.021 |
6.7 ± 0.028 | 9.376 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.171 ± 0.009 | 5.844 ± 0.019 |
3.87 ± 0.021 | 4.204 ± 0.017 |
4.884 ± 0.017 | 8.249 ± 0.026 |
5.573 ± 0.02 | 5.725 ± 0.015 |
1.122 ± 0.007 | 3.855 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
