
Fonsecaea pedrosoi CBS 271.37
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Fonsecaea; Fonsecaea pedrosoi
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
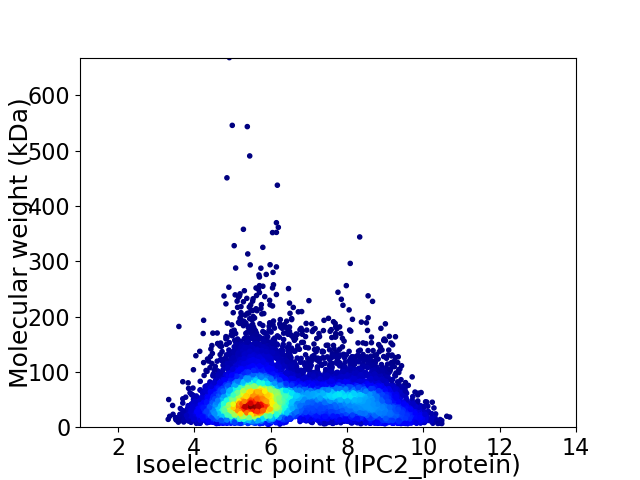
Virtual 2D-PAGE plot for 12525 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2GPT8|A0A0D2GPT8_9EURO Unplaced genomic scaffold supercont1.4 whole genome shotgun sequence OS=Fonsecaea pedrosoi CBS 271.37 OX=1442368 GN=Z517_07151 PE=3 SV=1
MM1 pKa = 7.2YY2 pKa = 10.32LLAGTAIAALATSCAASPGYY22 pKa = 10.46LKK24 pKa = 10.94LGLQRR29 pKa = 11.84RR30 pKa = 11.84APPEE34 pKa = 3.5GRR36 pKa = 11.84LIRR39 pKa = 11.84RR40 pKa = 11.84QDD42 pKa = 3.09SGGSVDD48 pKa = 5.78ALVTQNTNKK57 pKa = 10.35AEE59 pKa = 4.04YY60 pKa = 9.86LVNITVGTPPQDD72 pKa = 3.48LSVTLDD78 pKa = 3.4TGSSDD83 pKa = 3.13LWIPASSAKK92 pKa = 10.27LCQQGKK98 pKa = 9.66CDD100 pKa = 5.14LGAFDD105 pKa = 4.95PSKK108 pKa = 10.81SSTYY112 pKa = 11.0NVIEE116 pKa = 4.12EE117 pKa = 4.38GGFNITYY124 pKa = 9.51VQVGDD129 pKa = 3.94TDD131 pKa = 3.88AGDD134 pKa = 3.49WSSDD138 pKa = 3.4TITVGGSPSIEE149 pKa = 3.93DD150 pKa = 3.37QQLGVATALFDD161 pKa = 3.29QHH163 pKa = 7.62GVMGIGFDD171 pKa = 3.94TIEE174 pKa = 4.84ASNDD178 pKa = 3.48SPNGVYY184 pKa = 10.33PSVLDD189 pKa = 3.72NLKK192 pKa = 9.08STGIIDD198 pKa = 3.89RR199 pKa = 11.84KK200 pKa = 10.69AFSLYY205 pKa = 10.96LNDD208 pKa = 5.03LEE210 pKa = 4.33ATKK213 pKa = 10.72GAVIFGGIDD222 pKa = 3.15TTKK225 pKa = 10.69YY226 pKa = 10.78SGDD229 pKa = 3.62LVALPLQPGPDD240 pKa = 3.71GSVSEE245 pKa = 4.44YY246 pKa = 10.56FVTLTSVSFTDD257 pKa = 3.34STGQTTQLSPEE268 pKa = 4.79GYY270 pKa = 10.08AQAALLDD277 pKa = 4.09SGTTNTLLTNDD288 pKa = 3.47VLAPLALGLGAVVDD302 pKa = 4.57PSGEE306 pKa = 4.13AYY308 pKa = 10.42LVPCSLSSANGTIDD322 pKa = 3.39YY323 pKa = 11.06SFGGEE328 pKa = 4.12DD329 pKa = 4.51GITIQVPVSQVVGGQVYY346 pKa = 8.74TSSEE350 pKa = 3.9FDD352 pKa = 3.99DD353 pKa = 4.61PSGGCVLAFGSTAEE367 pKa = 4.25GAGASIFGDD376 pKa = 3.4SFLRR380 pKa = 11.84SAYY383 pKa = 10.44AVFDD387 pKa = 4.17IDD389 pKa = 3.55NQMAALAQAVEE400 pKa = 4.61NEE402 pKa = 4.61DD403 pKa = 3.64STSSIVVFPTGTTIPSATVTATATGTQLTEE433 pKa = 4.27VVSEE437 pKa = 4.13TDD439 pKa = 3.16IPTASIQSSTLVPGTPTFSLEE460 pKa = 4.23AASSTATTTGGSSSSSSSAAANNAVNAAIPTAALLGLGLAAAGMVGLL507 pKa = 4.75
MM1 pKa = 7.2YY2 pKa = 10.32LLAGTAIAALATSCAASPGYY22 pKa = 10.46LKK24 pKa = 10.94LGLQRR29 pKa = 11.84RR30 pKa = 11.84APPEE34 pKa = 3.5GRR36 pKa = 11.84LIRR39 pKa = 11.84RR40 pKa = 11.84QDD42 pKa = 3.09SGGSVDD48 pKa = 5.78ALVTQNTNKK57 pKa = 10.35AEE59 pKa = 4.04YY60 pKa = 9.86LVNITVGTPPQDD72 pKa = 3.48LSVTLDD78 pKa = 3.4TGSSDD83 pKa = 3.13LWIPASSAKK92 pKa = 10.27LCQQGKK98 pKa = 9.66CDD100 pKa = 5.14LGAFDD105 pKa = 4.95PSKK108 pKa = 10.81SSTYY112 pKa = 11.0NVIEE116 pKa = 4.12EE117 pKa = 4.38GGFNITYY124 pKa = 9.51VQVGDD129 pKa = 3.94TDD131 pKa = 3.88AGDD134 pKa = 3.49WSSDD138 pKa = 3.4TITVGGSPSIEE149 pKa = 3.93DD150 pKa = 3.37QQLGVATALFDD161 pKa = 3.29QHH163 pKa = 7.62GVMGIGFDD171 pKa = 3.94TIEE174 pKa = 4.84ASNDD178 pKa = 3.48SPNGVYY184 pKa = 10.33PSVLDD189 pKa = 3.72NLKK192 pKa = 9.08STGIIDD198 pKa = 3.89RR199 pKa = 11.84KK200 pKa = 10.69AFSLYY205 pKa = 10.96LNDD208 pKa = 5.03LEE210 pKa = 4.33ATKK213 pKa = 10.72GAVIFGGIDD222 pKa = 3.15TTKK225 pKa = 10.69YY226 pKa = 10.78SGDD229 pKa = 3.62LVALPLQPGPDD240 pKa = 3.71GSVSEE245 pKa = 4.44YY246 pKa = 10.56FVTLTSVSFTDD257 pKa = 3.34STGQTTQLSPEE268 pKa = 4.79GYY270 pKa = 10.08AQAALLDD277 pKa = 4.09SGTTNTLLTNDD288 pKa = 3.47VLAPLALGLGAVVDD302 pKa = 4.57PSGEE306 pKa = 4.13AYY308 pKa = 10.42LVPCSLSSANGTIDD322 pKa = 3.39YY323 pKa = 11.06SFGGEE328 pKa = 4.12DD329 pKa = 4.51GITIQVPVSQVVGGQVYY346 pKa = 8.74TSSEE350 pKa = 3.9FDD352 pKa = 3.99DD353 pKa = 4.61PSGGCVLAFGSTAEE367 pKa = 4.25GAGASIFGDD376 pKa = 3.4SFLRR380 pKa = 11.84SAYY383 pKa = 10.44AVFDD387 pKa = 4.17IDD389 pKa = 3.55NQMAALAQAVEE400 pKa = 4.61NEE402 pKa = 4.61DD403 pKa = 3.64STSSIVVFPTGTTIPSATVTATATGTQLTEE433 pKa = 4.27VVSEE437 pKa = 4.13TDD439 pKa = 3.16IPTASIQSSTLVPGTPTFSLEE460 pKa = 4.23AASSTATTTGGSSSSSSSAAANNAVNAAIPTAALLGLGLAAAGMVGLL507 pKa = 4.75
Molecular weight: 51.32 kDa
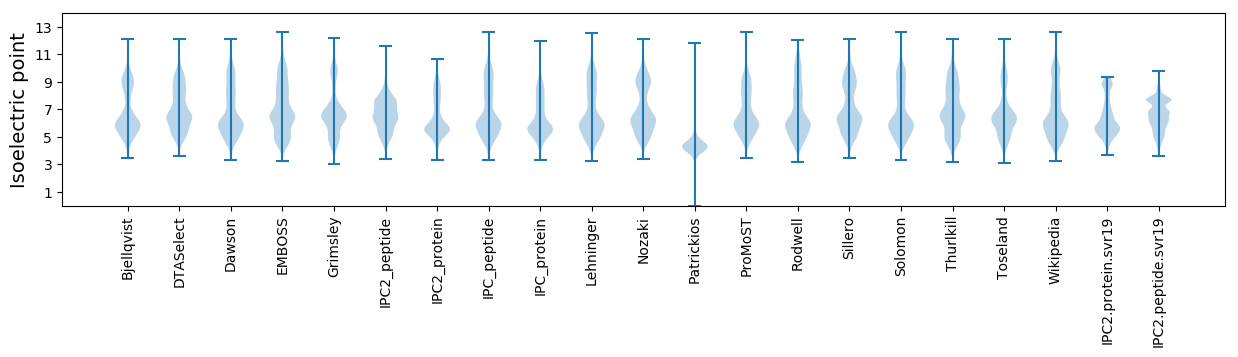
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2DUE2|A0A0D2DUE2_9EURO Unplaced genomic scaffold supercont1.3 whole genome shotgun sequence OS=Fonsecaea pedrosoi CBS 271.37 OX=1442368 GN=Z517_04416 PE=4 SV=1
MM1 pKa = 7.84ASTTFLQRR9 pKa = 11.84APKK12 pKa = 10.27SSQQEE17 pKa = 3.81LSSPNTHH24 pKa = 6.31KK25 pKa = 10.79RR26 pKa = 11.84LASLPPEE33 pKa = 3.78IHH35 pKa = 7.1HH36 pKa = 7.12IIADD40 pKa = 3.73HH41 pKa = 6.86LPYY44 pKa = 10.1PDD46 pKa = 4.87LLSLKK51 pKa = 9.81LTDD54 pKa = 3.57TYY56 pKa = 11.06FAALVGPKK64 pKa = 9.06LTVKK68 pKa = 10.54SRR70 pKa = 11.84VSWVQSRR77 pKa = 11.84YY78 pKa = 8.63AQRR81 pKa = 11.84LPVPTSTKK89 pKa = 10.34LSFRR93 pKa = 11.84TDD95 pKa = 3.2EE96 pKa = 4.53LFVANPEE103 pKa = 3.86VNAILRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84HH114 pKa = 5.9LEE116 pKa = 3.69CADD119 pKa = 3.47RR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 4.19PKK124 pKa = 10.33AALGFSAFTEE134 pKa = 3.72VDD136 pKa = 3.34RR137 pKa = 11.84NVRR140 pKa = 11.84RR141 pKa = 11.84LQGAQQLDD149 pKa = 3.76TTACLVTGAPNCPAIPNLDD168 pKa = 3.56TKK170 pKa = 8.13VQRR173 pKa = 11.84YY174 pKa = 8.55RR175 pKa = 11.84NSVVSKK181 pKa = 8.81LTAPVLRR188 pKa = 11.84TVAWLRR194 pKa = 11.84GLQRR198 pKa = 11.84SFRR201 pKa = 11.84VSLSCAVAEE210 pKa = 4.49LSHH213 pKa = 6.03TPRR216 pKa = 11.84VLWSWFLATLKK227 pKa = 10.61LAFIVVVASVACTVCGWRR245 pKa = 11.84PPPVAFTNPP254 pKa = 3.13
MM1 pKa = 7.84ASTTFLQRR9 pKa = 11.84APKK12 pKa = 10.27SSQQEE17 pKa = 3.81LSSPNTHH24 pKa = 6.31KK25 pKa = 10.79RR26 pKa = 11.84LASLPPEE33 pKa = 3.78IHH35 pKa = 7.1HH36 pKa = 7.12IIADD40 pKa = 3.73HH41 pKa = 6.86LPYY44 pKa = 10.1PDD46 pKa = 4.87LLSLKK51 pKa = 9.81LTDD54 pKa = 3.57TYY56 pKa = 11.06FAALVGPKK64 pKa = 9.06LTVKK68 pKa = 10.54SRR70 pKa = 11.84VSWVQSRR77 pKa = 11.84YY78 pKa = 8.63AQRR81 pKa = 11.84LPVPTSTKK89 pKa = 10.34LSFRR93 pKa = 11.84TDD95 pKa = 3.2EE96 pKa = 4.53LFVANPEE103 pKa = 3.86VNAILRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84HH114 pKa = 5.9LEE116 pKa = 3.69CADD119 pKa = 3.47RR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 4.19PKK124 pKa = 10.33AALGFSAFTEE134 pKa = 3.72VDD136 pKa = 3.34RR137 pKa = 11.84NVRR140 pKa = 11.84RR141 pKa = 11.84LQGAQQLDD149 pKa = 3.76TTACLVTGAPNCPAIPNLDD168 pKa = 3.56TKK170 pKa = 8.13VQRR173 pKa = 11.84YY174 pKa = 8.55RR175 pKa = 11.84NSVVSKK181 pKa = 8.81LTAPVLRR188 pKa = 11.84TVAWLRR194 pKa = 11.84GLQRR198 pKa = 11.84SFRR201 pKa = 11.84VSLSCAVAEE210 pKa = 4.49LSHH213 pKa = 6.03TPRR216 pKa = 11.84VLWSWFLATLKK227 pKa = 10.61LAFIVVVASVACTVCGWRR245 pKa = 11.84PPPVAFTNPP254 pKa = 3.13
Molecular weight: 28.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6141607 |
50 |
6165 |
490.3 |
54.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.629 ± 0.019 | 1.239 ± 0.008 |
5.737 ± 0.016 | 6.052 ± 0.021 |
3.74 ± 0.013 | 6.899 ± 0.02 |
2.468 ± 0.009 | 4.866 ± 0.014 |
4.668 ± 0.02 | 8.984 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.146 ± 0.007 | 3.541 ± 0.01 |
6.051 ± 0.024 | 4.093 ± 0.014 |
6.188 ± 0.021 | 8.097 ± 0.026 |
6.06 ± 0.016 | 6.279 ± 0.016 |
1.492 ± 0.008 | 2.77 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
